2NT3
 
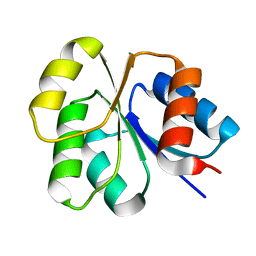 | | Receiver domain from Myxococcus xanthus social motility protein FrzS (Y102A Mutant) | | 分子名称: | Response regulator homolog | | 著者 | Fraser, J.S, Echols, N, Merlie, J.P, Zusman, D.R, Alber, T. | | 登録日 | 2006-11-06 | | 公開日 | 2007-03-13 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | An atypical receiver domain controls the dynamic polar localization of the Myxococcus xanthus social motility protein FrzS.
Mol.Microbiol., 65, 2007
|
|
3HTS
 
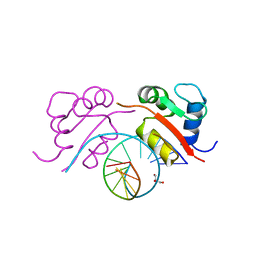 | | HEAT SHOCK TRANSCRIPTION FACTOR/DNA COMPLEX | | 分子名称: | 5'-D(*GP*GP*TP*TP*CP*TP*AP*GP*AP*AP*CP*C)-3', GLYCEROL, HEAT SHOCK TRANSCRIPTION FACTOR | | 著者 | Littlefield, O, Nelson, H.C.M. | | 登録日 | 1998-11-16 | | 公開日 | 1999-04-29 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | A new use for the 'wing' of the 'winged' helix-turn-helix motif in the HSF-DNA cocrystal.
Nat.Struct.Biol., 6, 1999
|
|
2MOA
 
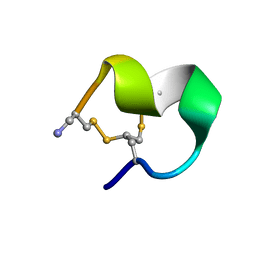 | |
1NF2
 
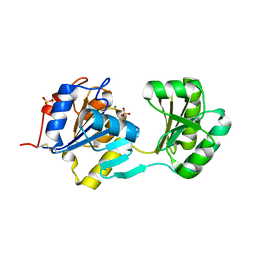 | |
1HRQ
 
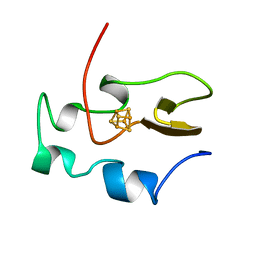 | | THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF THE REDUCED HIGH-POTENTIAL IRON-SULFUR PROTEIN FROM CHROMATIUM VINOSUM THROUGH NMR | | 分子名称: | HIGH POTENTIAL IRON SULFUR PROTEIN, IRON/SULFUR CLUSTER | | 著者 | Banci, L, Bertini, I, Dikiy, A, Kastrau, D.H.W, Luchinat, C, Sompornpisut, P. | | 登録日 | 1995-01-17 | | 公開日 | 1995-06-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The three-dimensional solution structure of the reduced high-potential iron-sulfur protein from Chromatium vinosum through NMR.
Biochemistry, 34, 1995
|
|
1HRR
 
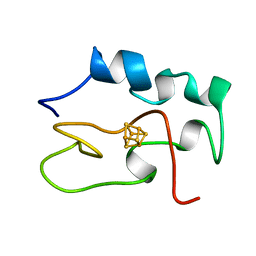 | | THE THREE DIMENSIONAL STRUCTURE OF THE REDUCED HIGH POTENTIAL IRON-SULFUR PROTEIN FROM CHROMATIUM VINOSUM THROUGH NMR | | 分子名称: | IRON/SULFUR CLUSTER, REDUCED HIGH POTENTIAL IRON SULFUR PROTEIN | | 著者 | Banci, L, Bertini, I, Dikiy, A, Kastrau, D.H.W, Luchinat, C, Sompornpisut, P. | | 登録日 | 1995-01-17 | | 公開日 | 1995-07-31 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The three-dimensional solution structure of the reduced high-potential iron-sulfur protein from Chromatium vinosum through NMR.
Biochemistry, 34, 1995
|
|
1XF6
 
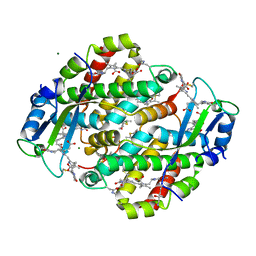 | | High resolution crystal structure of phycoerythrin 545 from the marine cryptophyte rhodomonas CS24 | | 分子名称: | 15,16-DIHYDROBILIVERDIN, B-phycoerythrin beta chain, CHLORIDE ION, ... | | 著者 | Doust, A.B, Marai, C.N.J, Harrop, S.J, Wilk, K.E, Curmi, P.M.G, Scholes, G.D. | | 登録日 | 2004-09-14 | | 公開日 | 2004-11-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Developing a structure-function model for the cryptophyte phycoerythrin 545 using ultrahigh resolution crystallography and ultrafast laser spectroscopy
J.Mol.Biol., 344, 2004
|
|
1XG0
 
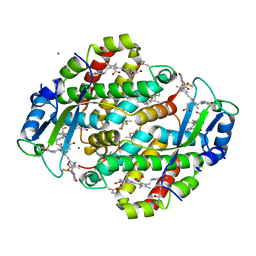 | | High resolution crystal structure of phycoerythrin 545 from the marine cryptophyte rhodomonas CS24 | | 分子名称: | 15,16-DIHYDROBILIVERDIN, B-phycoerythrin beta chain, CHLORIDE ION, ... | | 著者 | Doust, A.B, Marai, C.N.J, Harrop, S.J, Wilk, K.E, Curmi, P.M.G, Scholes, G.D. | | 登録日 | 2004-09-16 | | 公開日 | 2004-11-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (0.97 Å) | | 主引用文献 | Developing a structure-function model for the cryptophyte phycoerythrin 545 using ultrahigh resolution crystallography and ultrafast laser spectroscopy
J.Mol.Biol., 344, 2004
|
|
1MOL
 
 | |
1MDA
 
 | | CRYSTAL STRUCTURE OF AN ELECTRON-TRANSFER COMPLEX BETWEEN METHYLAMINE DEHYDROGENASE AND AMICYANIN | | 分子名称: | AMICYANIN, COPPER (II) ION, METHYLAMINE DEHYDROGENASE (HEAVY SUBUNIT), ... | | 著者 | Chen, L, Durley, R, Mathews, F.S. | | 登録日 | 1992-03-02 | | 公開日 | 1993-10-31 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of an electron-transfer complex between methylamine dehydrogenase and amicyanin.
Biochemistry, 31, 1992
|
|
1QGW
 
 | | CRYSTAL STRUCTURE OF PHYCOERYTHRIN 545 FROM THE MARINE CRYPTOPHYTE RHODOMONAS CS24 | | 分子名称: | 15,16-DIHYDROBILIVERDIN, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Harrop, S.J, Wilk, K.E, Hiller, R.G, Curmi, P.M.G. | | 登録日 | 1999-05-10 | | 公開日 | 1999-05-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Evolution of a light-harvesting protein by addition of new subunits and rearrangement of conserved elements: crystal structure of a cryptophyte phycoerythrin at 1.63-A resolution.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1G80
 
 | | NMR SOLUTION STRUCTURE OF D(GCGTACGC)2 | | 分子名称: | 5'-D(*GP*CP*GP*TP*AP*CP*GP*C)-3' | | 著者 | Isaacs, R.J, Spielmann, H.P. | | 登録日 | 2000-11-15 | | 公開日 | 2001-03-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Relationship of DNA structure to internal dynamics: correlation of helical parameters from NOE-based NMR solution structures of d(GCGTACGC)(2) and d(CGCTAGCG)(2) with (13)C order parameters implies conformational coupling in dinucleotide units.
J.Mol.Biol., 307, 2001
|
|
1G2G
 
 | | MINIMAL CONFORMATION OF THE ALPHA-CONOTOXIN IMI FOR THE ALPHA7 NEURONAL NICOTINIC ACETYLCHOLINE RECEPTOR RECOGNITION | | 分子名称: | ALPHA-CONOTOXIN IMI | | 著者 | Lamthanh, H, Jegou-Matheron, C, Servent, D, Menez, A, Lancelin, J.M. | | 登録日 | 2000-10-19 | | 公開日 | 2000-11-08 | | 最終更新日 | 2024-10-30 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Minimal conformation of the alpha-conotoxin ImI for the alpha7 neuronal nicotinic acetylcholine receptor recognition: correlated CD, NMR and binding studies.
FEBS Lett., 454, 1999
|
|
1G7Z
 
 | | NMR SOLUTION STRUCTURE OF D(CGCTAGCG)2 | | 分子名称: | 5'-D(*CP*GP*CP*TP*AP*GP*CP*G)-3' | | 著者 | Isaacs, R.J, Spielmann, H.P. | | 登録日 | 2000-11-15 | | 公開日 | 2001-03-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Relationship of DNA structure to internal dynamics: correlation of helical parameters from NOE-based NMR solution structures of d(GCGTACGC)(2) and d(CGCTAGCG)(2) with (13)C order parameters implies conformational coupling in dinucleotide units.
J.Mol.Biol., 307, 2001
|
|
