8RCT
 
 | |
2XL1
 
 | |
8PKL
 
 | |
8PEG
 
 | |
5LM7
 
 | | Crystal structure of the lambda N-Nus factor complex | | 分子名称: | 30S ribosomal protein S10, Antitermination protein N, N utilization substance protein B homolog, ... | | 著者 | Said, N, Santos, K, Weber, G, Wahl, M.C. | | 登録日 | 2016-07-29 | | 公開日 | 2017-04-05 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | Structural basis for lambda N-dependent processive transcription antitermination.
Nat Microbiol, 2, 2017
|
|
5LM9
 
 | | Structure of E. coli NusA | | 分子名称: | MAGNESIUM ION, SULFATE ION, Transcription termination/antitermination protein NusA | | 著者 | Said, N, Weber, G, Santos, K, Wahl, M.C. | | 登録日 | 2016-07-29 | | 公開日 | 2017-04-05 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.143 Å) | | 主引用文献 | Structural basis for lambda N-dependent processive transcription antitermination.
Nat Microbiol, 2, 2017
|
|
5MS0
 
 | |
7BL2
 
 | | pre-50S-ObgE particle state 1 | | 分子名称: | 23S ribosomal RNA, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | 著者 | Hilal, T, Nikolay, R, Schmidt, S, Spahn, C.M.T. | | 登録日 | 2021-01-18 | | 公開日 | 2021-05-12 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization.
Mol.Cell, 81, 2021
|
|
7BL5
 
 | | pre-50S-ObgE particle | | 分子名称: | 23S ribosomal RNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | 著者 | Hilal, T, Nikolay, R, Spahn, C.M.T, Schmidt, S. | | 登録日 | 2021-01-18 | | 公開日 | 2021-05-12 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization.
Mol.Cell, 81, 2021
|
|
7BL4
 
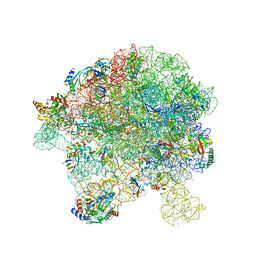 | | in vitro reconstituted 50S-ObgE-GMPPNP-RsfS particle | | 分子名称: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | 著者 | Hilal, T, Nikolay, R, Spahn, C.M.T. | | 登録日 | 2021-01-18 | | 公開日 | 2021-05-12 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.4 Å) | | 主引用文献 | Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization.
Mol.Cell, 81, 2021
|
|
7BL6
 
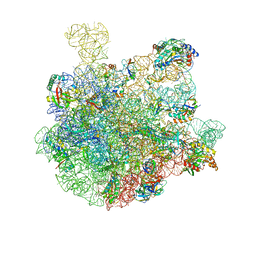 | | 50S-ObgE-GMPPNP particle | | 分子名称: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | 著者 | Hilal, T, Nikolay, R, Schmidt, S, Spahn, C.M.T. | | 登録日 | 2021-01-18 | | 公開日 | 2021-05-12 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization.
Mol.Cell, 81, 2021
|
|
7BL3
 
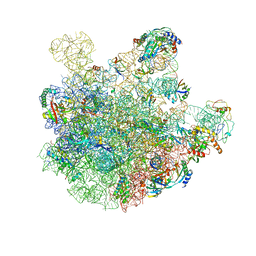 | | pre-50S-ObgE particle state 2 | | 分子名称: | 23S ribosomal RNA, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | 著者 | Hilal, T, Nikolay, R, Spahn, C.M.T. | | 登録日 | 2021-01-18 | | 公開日 | 2021-05-12 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization.
Mol.Cell, 81, 2021
|
|
7ADB
 
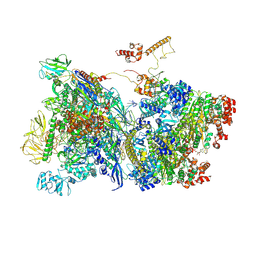 | | Transcription termination intermediate complex 1 delta NusG | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Said, N, Hilal, T, Loll, B, Wahl, C.M. | | 登録日 | 2020-09-14 | | 公開日 | 2020-11-04 | | 最終更新日 | 2021-02-03 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
7ADD
 
 | | Transcription termination intermediate complex IIIa | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Said, N, Hilal, T, Loll, B, Wahl, C.M. | | 登録日 | 2020-09-14 | | 公開日 | 2020-11-25 | | 最終更新日 | 2021-02-03 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
7ADE
 
 | | Transcription termination complex IVa | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Said, N, Hilal, T, Loll, B, Wahl, C.M. | | 登録日 | 2020-09-14 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
7ADC
 
 | | Transcription termination intermediate complex 3 delta NusG | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Said, N, Hilal, T, Loll, B, Wahl, C.M. | | 登録日 | 2020-09-14 | | 公開日 | 2020-11-25 | | 最終更新日 | 2021-02-03 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
5IFW
 
 | |
5IFS
 
 | | Quantitative interaction mapping reveals an extended ubiquitin regulatory domain in ASPL that disrupts functional p97 hexamers and induces cell death | | 分子名称: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | 著者 | Roske, Y, Arumughan, A, Heinemann, U, Wanker, E. | | 登録日 | 2016-02-26 | | 公開日 | 2016-10-26 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | Quantitative interaction mapping reveals an extended UBX domain in ASPL that disrupts functional p97 hexamers.
Nat Commun, 7, 2016
|
|
6SWA
 
 | |
7ZJX
 
 | | Rabbit 80S ribosome programmed with SECIS and SBP2 | | 分子名称: | 18S rRNA, 28S rRNA, 40S Ribosomal protein eS19, ... | | 著者 | Hilal, T, Simonovic, M, Spahn, C.M.T. | | 登録日 | 2022-04-12 | | 公開日 | 2022-09-07 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structure of the mammalian ribosome as it decodes the selenocysteine UGA codon.
Science, 376, 2022
|
|
7ZJW
 
 | | Rabbit 80S ribosome as it decodes the Sec-UGA codon | | 分子名称: | 18S rRNA, 28S rRNA, 40S Ribosomal protein eS19, ... | | 著者 | Hilal, T, Simonovic, M, Spahn, C.M.T. | | 登録日 | 2022-04-12 | | 公開日 | 2022-10-19 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structure of the mammalian ribosome as it decodes the selenocysteine UGA codon.
Science, 376, 2022
|
|
6TQO
 
 | | rrn anti-termination complex | | 分子名称: | 30S ribosomal protein S10, 30S ribosomal protein S4, DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Huang, Y.H, Wahl, M.C, Loll, B, Hilal, T, Said, N. | | 登録日 | 2019-12-17 | | 公開日 | 2020-08-05 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure-Based Mechanisms of a Molecular RNA Polymerase/Chaperone Machine Required for Ribosome Biosynthesis.
Mol.Cell, 79, 2020
|
|
6TQN
 
 | | rrn anti-termination complex without S4 | | 分子名称: | 30S ribosomal protein S10, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Huang, Y.H, Wahl, M.C, Loll, B, Hilal, T, Said, N. | | 登録日 | 2019-12-17 | | 公開日 | 2020-08-05 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure-Based Mechanisms of a Molecular RNA Polymerase/Chaperone Machine Required for Ribosome Biosynthesis.
Mol.Cell, 79, 2020
|
|
7ZN7
 
 | | Cryo-EM structure of RCMV-E E27 bound to human DDB1 (deltaBPB) and rat STAT2 CCD | | 分子名称: | B27a, DNA damage-binding protein 1, Signal transducer and activator of transcription, ... | | 著者 | Lauer, S, Spahn, C.M.T, Schwefel, D. | | 登録日 | 2022-04-20 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-03-15 | | 実験手法 | ELECTRON MICROSCOPY (3.78 Å) | | 主引用文献 | Structural mechanism of CRL4-instructed STAT2 degradation via a novel cytomegaloviral DCAF receptor.
Embo J., 42, 2023
|
|
7ZNN
 
 | | Cryo-EM structure of RCMV-E E27 bound to human DDB1 (deltaBPB) and full-length rat STAT2 | | 分子名称: | B27a, DNA damage-binding protein 1, Signal transducer and activator of transcription, ... | | 著者 | Lauer, S, Spahn, C.M.T, Schwefel, D. | | 登録日 | 2022-04-21 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-03-15 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Structural mechanism of CRL4-instructed STAT2 degradation via a novel cytomegaloviral DCAF receptor.
Embo J., 42, 2023
|
|
