4OX3
 
 | | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition | | 分子名称: | PHOSPHATE ION, Putative carboxypeptidase YodJ, ZINC ION | | 著者 | Hoyland, C.N, Aldridge, C, Cleverley, R.M, Sidiq, K, Duchene, M.C, Daniel, R.A, Vollmer, W, Lewis, R.J. | | 登録日 | 2014-02-04 | | 公開日 | 2014-06-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition.
Structure, 22, 2014
|
|
4OX5
 
 | | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | 著者 | Hoyland, C.N, Aldridge, C, Cleverley, R.M, Sidiq, K, Duchene, M.C, Daniel, R.A, Vollmer, W, Lewis, R.J. | | 登録日 | 2014-02-04 | | 公開日 | 2014-05-21 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition.
Structure, 22, 2014
|
|
4OXD
 
 | | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition | | 分子名称: | CHLORIDE ION, LYSINE, LdcB LD-carboxypeptidase, ... | | 著者 | Hoyland, C.N, Aldridge, C, Cleverley, R.M, Sidiq, K, Duchene, M.C, Daniel, R.A, Vollmer, W, Lewis, R.J. | | 登録日 | 2014-02-05 | | 公開日 | 2014-05-21 | | 最終更新日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition.
Structure, 22, 2014
|
|
3ZXJ
 
 | | Engineering the active site of a GH43 glycoside hydrolase generates a biotechnologically significant enzyme that displays both endo- xylanase and exo-arabinofuranosidase activity | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, HIAXHD3, ... | | 著者 | McKee, L.S, Pena, M.J, Rogowski, A, Jackson, A, Lewis, R.J, York, W.S, Krogh, K.B.R.M, Vikso-Nielsen, A, Skjot, M, Gilbert, H.J, Marles-Wright, J. | | 登録日 | 2011-08-11 | | 公開日 | 2012-04-18 | | 最終更新日 | 2012-05-02 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Introducing Endo-Xylanase Activity Into an Exo-Acting Arabinofuranosidase that Targets Side Chains.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZXK
 
 | | Engineering the active site of a GH43 glycoside hydrolase generates a biotechnologically significant enzyme that displays both endo- xylanase and exo-arabinofuranosidase activity | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIAXHD3, alpha-L-arabinofuranose-(1-2)-[beta-D-xylopyranose-(1-4)]beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | 著者 | McKee, L.S, Pena, M.J, Rogowski, A, Jackson, A, Lewis, R.J, York, W.S, Krogh, K.B.R.M, Vikso-Nielsen, A, Skjot, M, Gilbert, H.J, Marles-Wright, J. | | 登録日 | 2011-08-11 | | 公開日 | 2012-04-18 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | Introducing Endo-Xylanase Activity Into an Exo-Acting Arabinofuranosidase that Targets Side Chains.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZTB
 
 | | The bacterial stressosome: a modular system that has been adapted to control secondary messenger signaling | | 分子名称: | ANTI-SIGMA-FACTOR ANTAGONIST (STAS) DOMAIN PROTEIN, IODIDE ION | | 著者 | Quin, M.B, Berrisford, J.M, Newman, J.A, Basle, A, Lewis, R.J, Marles-Wright, J. | | 登録日 | 2011-07-06 | | 公開日 | 2012-02-22 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The Bacterial Stressosome: A Modular System that Has Been Adapted to Control Secondary Messenger Signaling.
Structure, 20, 2012
|
|
4B2O
 
 | | Crystal structure of Bacillus subtilis YmdB, a global regulator of late adaptive responses. | | 分子名称: | FE (II) ION, PHOSPHATE ION, YMDB PHOSPHODIESTERASE | | 著者 | Newman, J.A, Diethmaier, C, Kovacs, A.T, Rodrigues, C, Kuipers, O.P, Stulke, J, Lewis, R.J. | | 登録日 | 2012-07-17 | | 公開日 | 2013-07-24 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | The Ymdb Phosphodiesterase is a Global Regulator of Late Adaptive Responses in Bacillus Subtilis.
J.Bacteriol., 196, 2014
|
|
4A3R
 
 | | Crystal structure of Enolase from Bacillus subtilis. | | 分子名称: | CITRIC ACID, ENOLASE, SODIUM ION | | 著者 | Newman, J.A, Hewitt, L, Rodrigues, C, Solovyova, A.S, Harwood, C.R, Lewis, R.J. | | 登録日 | 2011-10-04 | | 公開日 | 2012-08-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Dissection of the Network of Interactions that Links RNA Processing with Glycolysis in the Bacillus Subtilis Degradosome.
J.Mol.Biol., 416, 2012
|
|
4A3S
 
 | | Crystal structure of PFK from Bacillus subtilis | | 分子名称: | 6-PHOSPHOFRUCTOKINASE | | 著者 | Newman, J.A, Hewitt, L, Rodrigues, C, Solovyova, A.S, Harwood, C.R, Lewis, R.J. | | 登録日 | 2011-10-04 | | 公開日 | 2012-08-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Dissection of the Network of Interactions that Links RNA Processing with Glycolysis in the Bacillus Subtilis Degradosome.
J.Mol.Biol., 416, 2012
|
|
4AXO
 
 | | Structure of the Clostridium difficile EutQ protein | | 分子名称: | ETHANOLAMINE UTILIZATION PROTEIN, MAGNESIUM ION | | 著者 | Pitts, A.C, Tuck, L.R, Faulds-Pain, A, Lewis, R.J, Marles-Wright, J. | | 登録日 | 2012-06-13 | | 公開日 | 2012-06-27 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Structural Insight Into the Clostridium Difficile Ethanolamine Utilisation Microcompartment.
Plos One, 7, 2012
|
|
4AXJ
 
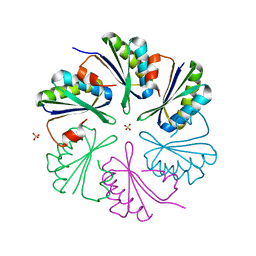 | | Structure of the Clostridium difficile EutM protein | | 分子名称: | ETHANOLAMINE CARBOXYSOME STRUCTURAL PROTEIN, SULFATE ION | | 著者 | Pitts, A.C, Tuck, L.R, Faulds-Pain, A, Lewis, R.J, Marles-Wright, J. | | 登録日 | 2012-06-13 | | 公開日 | 2012-06-20 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Structural Insight Into the Clostridium Difficile Ethanolamine Utilisation Microcompartment.
Plos One, 7, 2012
|
|
4AXI
 
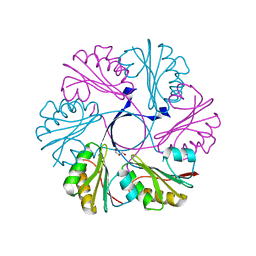 | | Structure of the Clostridium difficile EutS protein | | 分子名称: | ETHANOLAMINE CARBOXYSOME STRUCTURAL PROTEIN, GLYCEROL | | 著者 | Pitts, A.C, Tuck, L.R, Faulds-Pain, A, Lewis, R.J, Marles-Wright, J. | | 登録日 | 2012-06-13 | | 公開日 | 2012-06-27 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Structural Insight Into the Clostridium Difficile Ethanolamine Utilisation Microcompartment.
Plos One, 7, 2012
|
|
3ZQ4
 
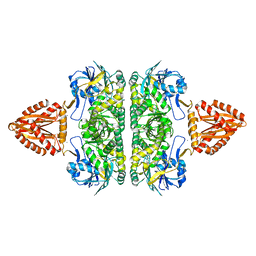 | | Unusual, dual endo- and exo-nuclease activity in the degradosome explained by crystal structure analysis of RNase J1 | | 分子名称: | CALCIUM ION, RIBONUCLEASE J 1, ZINC ION | | 著者 | Newman, J.A, Hewitt, L, Rodrigues, C, Solovyova, A, Harwood, C.R, Lewis, R.J. | | 登録日 | 2011-06-07 | | 公開日 | 2011-09-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Unusual, Dual Endo- and Exonuclease Activity in the Degradosome Explained by Crystal Structure Analysis of Rnase J1.
Structure, 19, 2011
|
|
3ZXL
 
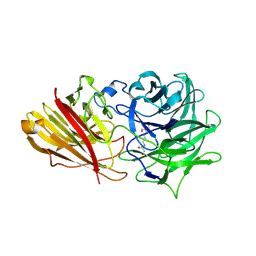 | | Engineering the active site of a GH43 glycoside hydrolase generates a biotechnologically significant enzyme that displays both endo- xylanase and exo-arabinofuranosidase activity | | 分子名称: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HIAXHD3 | | 著者 | McKee, L.S, Pena, M.J, Rogowski, A, Jackson, A, Lewis, R.J, York, W.S, Krogh, K.B.R.M, Vikso-Nielsen, A, Skjot, M, Gilbert, H.J, Marles-Wright, J. | | 登録日 | 2011-08-11 | | 公開日 | 2012-04-18 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.871 Å) | | 主引用文献 | Introducing Endo-Xylanase Activity Into an Exo-Acting Arabinofuranosidase that Targets Side Chains.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZXN
 
 | | Moorella thermoacetica RsbS S58E | | 分子名称: | ANTI-SIGMA-FACTOR ANTAGONIST (STAS) DOMAIN PROTEIN, THIOCYANATE ION | | 著者 | Quin, M.B, Berrisford, J.M, Newman, J.A, Basle, A, Lewis, R.J, Marles-Wright, J. | | 登録日 | 2011-08-12 | | 公開日 | 2012-02-22 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The Bacterial Stressosome: A Modular System that Has Been Adapted to Control Secondary Messenger Signaling.
Structure, 20, 2012
|
|
1TTK
 
 | | NMR solution structure of omega-conotoxin MVIIA, a N-type calcium channel blocker | | 分子名称: | Omega-conotoxin MVIIa | | 著者 | Adams, D.J, Smith, A.B, Schroeder, C.I, Yasuda, T, Lewis, R.J. | | 登録日 | 2004-06-22 | | 公開日 | 2004-07-06 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | omega-conotoxin CVID inhibits a pharmacologically distinct voltage-sensitive calcium channel associated with transmitter release from preganglionic nerve terminals
J.Biol.Chem., 278, 2003
|
|
1TT3
 
 | | NMR soulution structure of omega-conotoxin [K10]MVIIA | | 分子名称: | Omega-conotoxin MVIIa | | 著者 | Adams, D.J, Smith, A.B, Schroeder, C.I, Yasuda, T, Lewis, R.J. | | 登録日 | 2004-06-21 | | 公開日 | 2004-07-06 | | 最終更新日 | 2021-11-10 | | 実験手法 | SOLUTION NMR | | 主引用文献 | omega-conotoxin CVID inhibits a pharmacologically distinct voltage-sensitive calcium channel associated with transmitter release from preganglionic nerve terminals
J.Biol.Chem., 278, 2003
|
|
1FSE
 
 | | CRYSTAL STRUCTURE OF THE BACILLUS SUBTILIS REGULATORY PROTEIN GERE | | 分子名称: | GERE, GLYCEROL, SULFATE ION | | 著者 | Ducros, V.M.-A, Lewis, R.J, Verma, C.S, Dodson, E.J, Leonard, G, Turkenburg, J.P, Murshudov, G.N, Wilkinson, A.J, Brannigan, J.A. | | 登録日 | 2000-09-08 | | 公開日 | 2001-03-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Crystal structure of GerE, the ultimate transcriptional regulator of spore formation in Bacillus subtilis.
J.Mol.Biol., 306, 2001
|
|
1TTL
 
 | | Omega-conotoxin GVIA, a N-type calcium channel blocker | | 分子名称: | Omega-conotoxin GVIA | | 著者 | Mould, J, Yasuda, T, Schroeder, C.I, Beedle, A.M, Doering, C.J, Zamponi, G.W, Adams, D.J, Lewis, R.J. | | 登録日 | 2004-06-23 | | 公開日 | 2004-07-13 | | 最終更新日 | 2011-07-13 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The alpha2delta auxiliary subunit reduces affinity of omega-conotoxins for recombinant N-type (Cav2.2) calcium channels
J.Biol.Chem., 279, 2004
|
|
1TR6
 
 | | NMR solution structure of omega-conotoxin [K10]GVIA, a cyclic cysteine knot peptide | | 分子名称: | Omega-conotoxin GVIA | | 著者 | Mould, J, Yasuda, T, Schroeder, C.I, Beedle, A.M, Doering, C.J, Zamponi, G.W, Adams, D.J, Lewis, R.J. | | 登録日 | 2004-06-21 | | 公開日 | 2004-07-13 | | 最終更新日 | 2011-10-05 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The alpha2delta auxiliary subunit reduces affinity of omega-conotoxins for recombinant N-type (Cav2.2) calcium channels
J.Biol.Chem., 279, 2004
|
|
1W53
 
 | | Kinase recruitment domain of the stress phosphatase RsbU | | 分子名称: | GLYCEROL, PHOSPHOSERINE PHOSPHATASE RSBU, XENON | | 著者 | Delumeau, O, Dutta, S, Brigulla, M, Kuhnke, G, Hardwick, S.W, Voelker, U, Yudkin, M.D, Lewis, R.J. | | 登録日 | 2004-08-05 | | 公開日 | 2004-08-05 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Functional and Structural Characterization of Rsbu, a Stress Signaling Protein Phosphatase 2C
J.Biol.Chem., 279, 2004
|
|
1RMK
 
 | | Solution structure of conotoxin MrVIB | | 分子名称: | Mu-O-conotoxin MrVIB | | 著者 | Daly, N.L, Ekberg, J.A, Thomas, L, Adams, D.J, Lewis, R.J, Craik, D.J. | | 登録日 | 2003-11-28 | | 公開日 | 2004-09-07 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structures of muO-conotoxins from Conus marmoreus. Inhibitors of tetrodotoxin (TTX)-sensitive and TTX-resistant sodium channels in mammalian sensory neurons
J.Biol.Chem., 279, 2004
|
|
1ONT
 
 | | NMDA RECEPTOR ANTAGONIST, CONANTOKIN-T, NMR, 17 STRUCTURES | | 分子名称: | CONANTOKIN-T | | 著者 | Skjaerbaek, N, Nielsen, K.J, Lewis, R.J, Alewood, P.F, Craik, D.J. | | 登録日 | 1996-08-27 | | 公開日 | 1997-09-04 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Determination of the solution structures of conantokin-G and conantokin-T by CD and NMR spectroscopy.
J.Biol.Chem., 272, 1997
|
|
1ONU
 
 | | NMDA RECEPTOR ANTAGONIST, CONANTOKIN-G, NMR, 17 STRUCTURES | | 分子名称: | CONANTOKIN-G | | 著者 | Skjaerbaek, N, Nielsen, K.J, Lewis, R.J, Alewood, P.F, Craik, D.J. | | 登録日 | 1996-08-27 | | 公開日 | 1997-09-04 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Determination of the solution structures of conantokin-G and conantokin-T by CD and NMR spectroscopy.
J.Biol.Chem., 272, 1997
|
|
1R9I
 
 | | NMR Solution Structure of PIIIA toxin, NMR, 20 structures | | 分子名称: | Mu-conotoxin PIIIA | | 著者 | Nielsen, K.J, Watson, M, Adams, D.J, Hammarstrom, A.K, Gage, P.W, Hill, J.M, Craik, D.J, Thomas, L, Adams, D, Alewood, P.F, Lewis, R.J. | | 登録日 | 2003-10-30 | | 公開日 | 2003-11-18 | | 最終更新日 | 2019-12-25 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of mu-conotoxin PIIIA, a preferential inhibitor of persistent tetrodotoxin-sensitive sodium channels
J.Biol.Chem., 277, 2002
|
|
