1FXD
 
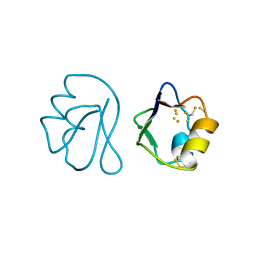 | | REFINED CRYSTAL STRUCTURE OF FERREDOXIN II FROM DESULFOVIBRIO GIGAS AT 1.7 ANGSTROMS | | 分子名称: | FE3-S4 CLUSTER, FERREDOXIN II | | 著者 | Kissinger, C.R, Sieker, L.C, Adman, E.T, Jensen, L.H. | | 登録日 | 1991-04-08 | | 公開日 | 1993-04-15 | | 最終更新日 | 2017-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Refined crystal structure of ferredoxin II from Desulfovibrio gigas at 1.7 A.
J.Mol.Biol., 219, 1991
|
|
6RA6
 
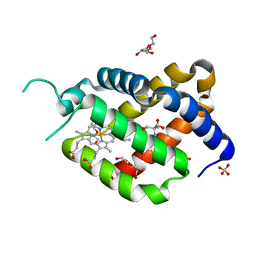 | | Ferric murine neuroglobin Gly-loop44-47/F106A mutant | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Exertier, C, Freda, I, Montemiglio, L.C, Savino, C, Vallone, B. | | 登録日 | 2019-04-05 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Lack of orientation selectivity of the heme insertion in murine neuroglobin revealed by resonance Raman spectroscopy.
Febs J., 287, 2020
|
|
6S53
 
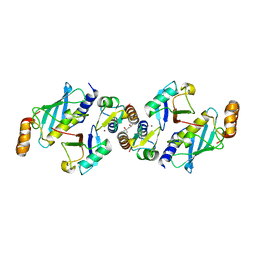 | | Crystal structure of TRIM21 RING domain in complex with an isopeptide-linked Ube2N~ubiquitin conjugate | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, E3 ubiquitin-protein ligase TRIM21, Polyubiquitin-C, ... | | 著者 | Kiss, L, Boland, A, Neuhaus, D, James, L.C. | | 登録日 | 2019-06-30 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A tri-ionic anchor mechanism drives Ube2N-specific recruitment and K63-chain ubiquitination in TRIM ligases.
Nat Commun, 10, 2019
|
|
6RSS
 
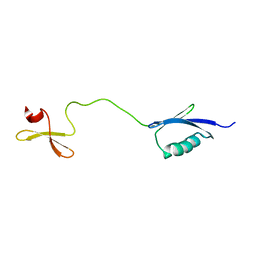 | | Solution structure of the fourth WW domain of WWP2 with GB1-tag | | 分子名称: | NEDD4-like E3 ubiquitin-protein ligase WWP2 | | 著者 | Wahl, L.C, Watt, J.E, Tolchard, J, Blumenschein, T.M.A, Chantry, A. | | 登録日 | 2019-05-22 | | 公開日 | 2019-10-09 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Smad7 Binds Differently to Individual and Tandem WW3 and WW4 Domains of WWP2 Ubiquitin Ligase Isoforms.
Int J Mol Sci, 20, 2019
|
|
8A58
 
 | |
6R6Q
 
 | |
6TYW
 
 | |
6TYZ
 
 | |
6TYV
 
 | |
7KU0
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 138 (yellow) structure | | 分子名称: | Chymotrypsinogen A, SULFATE ION | | 著者 | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | 登録日 | 2020-11-24 | | 公開日 | 2021-03-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU2
 
 | | Data clustering and dynamics of chymotrypsinogen clulster 140 (structure) | | 分子名称: | Chymotrypsinogen A, SULFATE ION | | 著者 | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | 登録日 | 2020-11-24 | | 公開日 | 2021-03-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.185 Å) | | 主引用文献 | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU1
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 139 (green) structure | | 分子名称: | Chymotrypsinogen A, SULFATE ION | | 著者 | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | 登録日 | 2020-11-24 | | 公開日 | 2021-03-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU3
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 141 (cyan) structure | | 分子名称: | Chymotrypsinogen A, SULFATE ION | | 著者 | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | 登録日 | 2020-11-24 | | 公開日 | 2021-03-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KTZ
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 131 (purple) structure | | 分子名称: | Chymotrypsinogen A, SULFATE ION | | 著者 | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | 登録日 | 2020-11-24 | | 公開日 | 2021-03-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KTY
 
 | | Data clustering and dynamics of chymotrypsinogen average structure | | 分子名称: | Chymotrypsinogen A, SULFATE ION | | 著者 | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Shi, W, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | 登録日 | 2020-11-24 | | 公開日 | 2021-03-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7JYZ
 
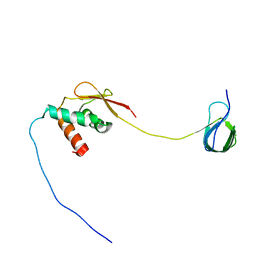 | | Solution NMR structure and dynamics of human Brd3 ET in complex with MLV IN CTD | | 分子名称: | Bromodomain-containing protein 3, Integrase | | 著者 | Aiyer, S, Liu, G, Swapna, G.V.T, Hao, J, Ma, L.C, Roth, M.J, Montelione, G.T. | | 登録日 | 2020-09-01 | | 公開日 | 2021-06-23 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A common binding motif in the ET domain of BRD3 forms polymorphic structural interfaces with host and viral proteins.
Structure, 29, 2021
|
|
1KEM
 
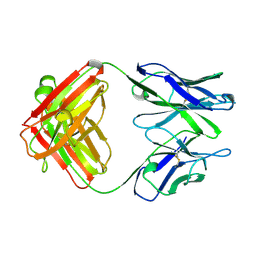 | |
1FX1
 
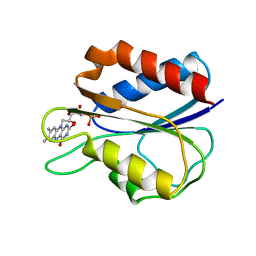 | |
1KEL
 
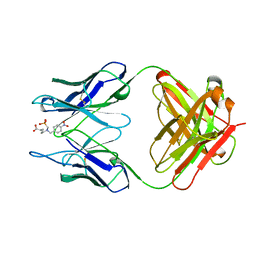 | |
6IDW
 
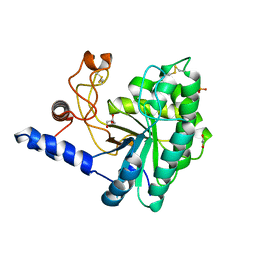 | | GH6 Orpinomyces sp. Y102 enzyme | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, GLYCEROL, ... | | 著者 | Tsai, L.C, Huang, H.C. | | 登録日 | 2018-09-11 | | 公開日 | 2018-10-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.78 Å) | | 主引用文献 | Crystal structures of the GH6 Orpinomyces sp. Y102 CelC7 enzyme with exo and endo activity and its complex with cellobiose.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6H09
 
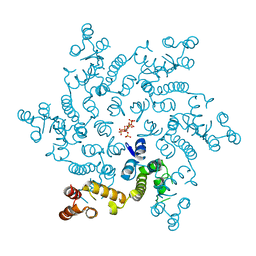 | | HIV capsid hexamer with IP6 ligand | | 分子名称: | Gag polyprotein, INOSITOL HEXAKISPHOSPHATE | | 著者 | James, L.C. | | 登録日 | 2018-07-06 | | 公開日 | 2018-08-15 | | 最終更新日 | 2020-10-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | IP6 is an HIV pocket factor that prevents capsid collapse and promotes DNA synthesis.
Elife, 7, 2018
|
|
6H5I
 
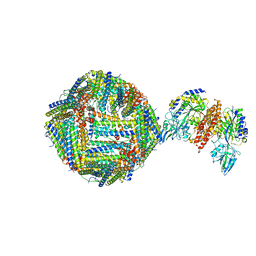 | | Single Particle Cryo-EM map of human Transferrin receptor 1 - H-Ferritin complex. | | 分子名称: | Ferritin heavy chain, Transferrin receptor protein 1 | | 著者 | Testi, C, Montemiglio, L.C, Vallone, B, Des Georges, A, Boffi, A, Mancia, F, Baiocco, P, Savino, C. | | 登録日 | 2018-07-24 | | 公開日 | 2019-03-27 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Cryo-EM structure of the human ferritin-transferrin receptor 1 complex.
Nat Commun, 10, 2019
|
|
6GL1
 
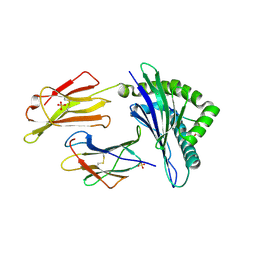 | | HLA-E*01:03 in complex with the HIV epitope, RL9HIV | | 分子名称: | ARG-MET-TYR-SER-PRO-THR-SER-ILE-LEU, Beta-2-microglobulin, MHC class I antigen, ... | | 著者 | Walters, L.C, Gillespie, G.M, McMichael, A.J, Rozbesky, D, Jones, E.Y, Harlos, K. | | 登録日 | 2018-05-22 | | 公開日 | 2018-08-08 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.623 Å) | | 主引用文献 | Pathogen-derived HLA-E bound epitopes reveal broad primary anchor pocket tolerability and conformationally malleable peptide binding.
Nat Commun, 9, 2018
|
|
3CS6
 
 | | Structure-based design of a superagonist ligand for the vitamin D nuclear receptor | | 分子名称: | (1S,3R,5Z,7E,14beta,17alpha,23R)-23-(2-hydroxy-2-methylpropyl)-20,24-epoxy-9,10-secochola-5,7,10-triene-1,3-diol, Vitamin D3 receptor | | 著者 | Hourai, S, Rodriguez, L.C, Antony, P, Reina-San-Martin, B, Ciesielski, P, Magnier, B.C, Schoonjans, K, Mourino, A, Rochel, N, Moras, D. | | 登録日 | 2008-04-09 | | 公開日 | 2008-05-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure-based design of a superagonist ligand for the vitamin d nuclear receptor.
Chem.Biol., 15, 2008
|
|
6I40
 
 | | Crystal structure of murine neuroglobin bound to CO at 15K under illumination using optical fiber | | 分子名称: | ACETATE ION, CARBON MONOXIDE, FORMIC ACID, ... | | 著者 | Savino, C, Montemiglio, L.C, Ardiccioni, C, Exertier, C, Vallone, B. | | 登録日 | 2018-11-08 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Ligand pathways in neuroglobin revealed by low-temperature photodissociation and docking experiments.
Iucrj, 6, 2019
|
|
