5ENF
 
 | | Crystal structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with fragment-4 N10142 (SGC - Diamond I04-1 fragment screening) | | 分子名称: | 1,2-ETHANEDIOL, 5-azanyl-2-(2-methylpropyl)-1,3-oxazole-4-carbonitrile, PH-interacting protein | | 著者 | Krojer, T, Talon, R, Collins, P, Bradley, A, Cox, O, Amin, J, Szykowska, A, Burgess-Brown, N, Spencer, J, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F, Structural Genomics Consortium (SGC) | | 登録日 | 2015-11-09 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.37 Å) | | 主引用文献 | A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain.
Chem Sci, 7, 2016
|
|
5ENH
 
 | | Crystal structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with compound-12 N11528 (SGC - Diamond I04-1 fragment screening) | | 分子名称: | PH-interacting protein, ~{N}-[(2,6-dimethoxyphenyl)methyl]ethanamide | | 著者 | Krojer, T, Talon, R, Collins, P, Bradley, A, Cox, O, Szykowska, A, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F, Structural Genomics Consortium (SGC) | | 登録日 | 2015-11-09 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain.
Chem Sci, 7, 2016
|
|
5ENB
 
 | | Crystal structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with o-Tolylthiourea (SGC - Diamond I04-1 fragment screening) | | 分子名称: | 1,2-ETHANEDIOL, 1-(2-methylphenyl)thiourea, PH-interacting protein | | 著者 | Krojer, T, Talon, R, Collins, P, Bradley, A, Cox, O, Szykowska, A, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F, Structural Genomics Consortium (SGC) | | 登録日 | 2015-11-09 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain.
Chem Sci, 7, 2016
|
|
5ENI
 
 | | Crystal structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with compound-13 N11537 (SGC - Diamond I04-1 fragment screening) | | 分子名称: | PH-interacting protein, ~{N}-[[2,6-bis(chloranyl)phenyl]methyl]-2-oxidanyl-ethanamide | | 著者 | Krojer, T, Talon, R, Collins, P, Bradley, A, Cox, O, Szykowska, A, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F, Structural Genomics Consortium (SGC) | | 登録日 | 2015-11-09 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain.
Chem Sci, 7, 2016
|
|
5ENJ
 
 | | Crystal structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with compound-14 N11530 (SGC - Diamond I04-1 fragment screening) | | 分子名称: | MAGNESIUM ION, PH-interacting protein, ~{N}-[[2,6-bis(chloranyl)phenyl]methyl]-~{N}-methyl-ethanamide | | 著者 | Krojer, T, Talon, R, Collins, P, Bradley, A, Cox, O, Szykowska, A, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F, Structural Genomics Consortium (SGC) | | 登録日 | 2015-11-09 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain.
Chem Sci, 7, 2016
|
|
5ENE
 
 | | Crystal structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with 5-Amino-2-benzyl-1,3-oxazole-4-carbonitrile (SGC - Diamond I04-1 fragment screening) | | 分子名称: | 5-azanyl-2-(phenylmethyl)-1,3-oxazole-4-carbonitrile, PH-interacting protein | | 著者 | Krojer, T, Talon, R, Collins, P, Bradley, A, Cox, O, Amin, J, Szykowska, A, Burgess-Brown, N, Spencer, J, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F, Structural Genomics Consortium (SGC) | | 登録日 | 2015-11-09 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain.
Chem Sci, 7, 2016
|
|
5ENC
 
 | | Crystal structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with N-(2,6-Dichlorobenzyl)acetamide (SGC - Diamond I04-1 fragment screening) | | 分子名称: | 1,2-ETHANEDIOL, PH-interacting protein, ~{N}-[[2,6-bis(chloranyl)phenyl]methyl]ethanamide | | 著者 | Krojer, T, Talon, R, Collins, P, Bradley, A, Cox, O, Szykowska, A, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F, Structural Genomics Consortium (SGC) | | 登録日 | 2015-11-09 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain.
Chem Sci, 7, 2016
|
|
5F5C
 
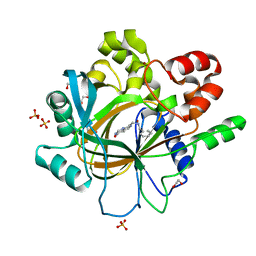 | | Crystal Structure of human JMJD2D complexed with KDOPP7 | | 分子名称: | 1,2-ETHANEDIOL, 8-[[(phenylmethyl)amino]methyl]-1~{H}-pyrido[3,4-d]pyrimidin-4-one, Lysine-specific demethylase 4D, ... | | 著者 | Krojer, T, Vollmar, M, Crawley, L, Bradley, A.R, Szykowska, A, Ruda, G.F, Yang, H, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, Oppermann, U, von Delft, F, Structural Genomics Consortium (SGC) | | 登録日 | 2015-12-04 | | 公開日 | 2015-12-30 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
5F5A
 
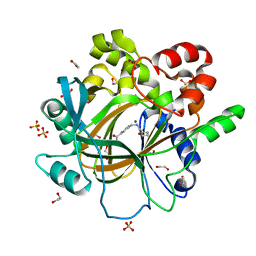 | | Crystal Structure of human JMJD2D complexed with KDOAM16 | | 分子名称: | 1,2-ETHANEDIOL, 2-[(furan-2-ylmethylamino)methyl]pyridine-4-carboxylic acid, Lysine-specific demethylase 4D, ... | | 著者 | Krojer, T, Vollmar, M, Crawley, L, Bradley, A.R, Szykowska, A, Ruda, G.F, Yang, H, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, Oppermann, U, von Delft, F, Structural Genomics Consortium (SGC) | | 登録日 | 2015-12-04 | | 公開日 | 2015-12-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
5F5I
 
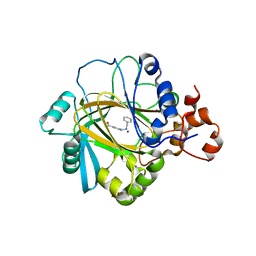 | | Crystal Structure of human JMJD2A complexed with KDOOA011340 | | 分子名称: | 2-[[(phenylmethyl)amino]methyl]pyridine-4-carboxylic acid, Lysine-specific demethylase 4A, NICKEL (II) ION, ... | | 著者 | Krojer, T, Vollmar, M, Crawley, L, Szykowska, A, Gileadi, C, Johansson, C, England, K, Yang, H, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, Oppermann, U, von Delft, F, Structural Genomics Consortium (SGC) | | 登録日 | 2015-12-04 | | 公開日 | 2015-12-30 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.63 Å) | | 主引用文献 | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
4CRI
 
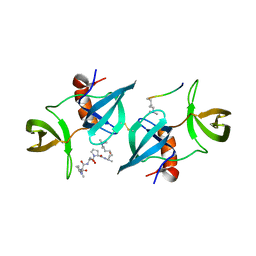 | | Crystal Structure of 53BP1 tandem tudor domains in complex with methylated K810 Rb peptide | | 分子名称: | RB1 PROTEIN, TUMOR SUPPRESSOR P53-BINDING PROTEIN 1 | | 著者 | Krojer, T, Johansson, C, Gileadi, C, Fedorov, O, Carr, S, La Thangue, N.B, Vollmar, M, Crawley, L, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Oppermann, U. | | 登録日 | 2014-02-26 | | 公開日 | 2014-08-06 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Lysine Methylation-Dependent Binding of 53BP1 to the Prb Tumor Suppressor.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3SOM
 
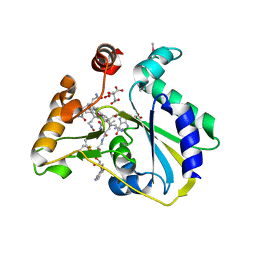 | | crystal structure of human MMACHC | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-ETHANETHIOL, 5'-DEOXYADENOSINE, ... | | 著者 | Krojer, T, Froese, D.S, von Delft, F, Muniz, J.R, Gileadi, C, Vollmar, M, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Gravel, R.A, Yue, W.W, Oppermann, U, Structural Genomics Consortium (SGC) | | 登録日 | 2011-06-30 | | 公開日 | 2011-07-27 | | 最終更新日 | 2015-04-15 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure of MMACHC reveals an arginine-rich pocket and a domain-swapped dimer for its B12 processing function.
Biochemistry, 51, 2012
|
|
5APA
 
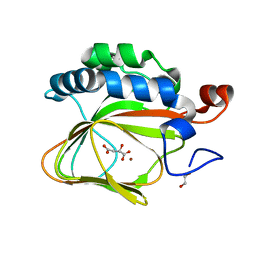 | | Crystal structure of human aspartate beta-hydroxylase isoform a | | 分子名称: | (2S)-2-hydroxybutanedioic acid, 1,2-ETHANEDIOL, ASPARTYL/ASPARAGINYL BETA-HYDROXYLASE, ... | | 著者 | Krojer, T, Kochan, G, Pfeffer, I, McDonough, M.A, Pilka, E, Hozjan, V, Allerston, C, Muniz, J.R, Chaikuad, A, Gileadi, O, Kavanagh, K, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Oppermann, U. | | 登録日 | 2015-09-15 | | 公開日 | 2015-09-23 | | 最終更新日 | 2019-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Aspartate/asparagine-beta-hydroxylase crystal structures reveal an unexpected epidermal growth factor-like domain substrate disulfide pattern.
Nat Commun, 10, 2019
|
|
2XDV
 
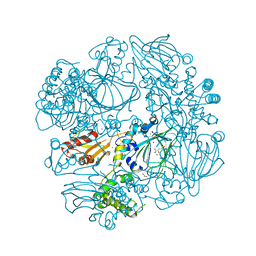 | | Crystal Structure of the Catalytic Domain of FLJ14393 | | 分子名称: | 1,2-ETHANEDIOL, CADMIUM ION, MANGANESE (II) ION, ... | | 著者 | Krojer, T, Muniz, J.R.C, Ng, S.S, Pilka, E, Guo, K, Pike, A.C.W, Filippakopoulos, P, Knapp, S, Kavanagh, K.L, Gileadi, O, Bunkoczi, G, Yue, W.W, Niesen, F, Sobott, F, Fedorov, O, Savitsky, P, Kochan, G, Daniel, M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Oppermann, U. | | 登録日 | 2010-05-07 | | 公開日 | 2010-05-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.57 Å) | | 主引用文献 | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
8C8A
 
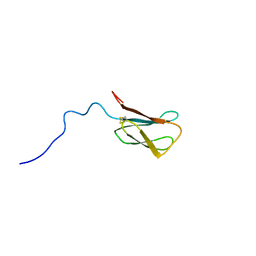 | | The NMR structure of the MAX28 effector from Magnaporthe oryzae | | 分子名称: | But2 domain-containing protein | | 著者 | Lahfa, M, Padilla, A, de Guillen, K, Kroj, T, Roumestand, C, Barthe, P. | | 登録日 | 2023-01-19 | | 公開日 | 2024-02-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | 1H, 15N Backbone and side-chain NMR assignments for a MAX effector from Magnaporthe oryzae
To Be Published
|
|
2MYW
 
 | |
2MYV
 
 | |
7ZKD
 
 | | The NMR structure of the MAX47 effector from Magnaporthe Oryzae | | 分子名称: | MAX effector protein | | 著者 | Lahfa, M, Padilla, A, de Guillen, K, Pissarra, J, Raji, M, Cesari, S, Kroj, T, Gladieux, P, Roumestand, C, Barthe, P. | | 登録日 | 2022-04-12 | | 公開日 | 2023-04-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | 1 H, 13 C, 15 N backbone and side-chain NMR assignments for three MAX effectors from Magnaporthe oryzae.
Biomol.Nmr Assign., 16, 2022
|
|
7ZJY
 
 | | The NMR structure of the MAX67 effector from Magnaporthe Oryzae | | 分子名称: | MAX effector protein | | 著者 | Lahfa, M, Padilla, A, de Guillen, K, Pissarra, J, Raji, M, Cesari, S, Kroj, T, Gladieux, P, Roumestand, C, Barthe, P. | | 登録日 | 2022-04-12 | | 公開日 | 2023-04-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | 1 H, 13 C, 15 N backbone and side-chain NMR assignments for three MAX effectors from Magnaporthe oryzae.
Biomol.Nmr Assign., 16, 2022
|
|
7ZK0
 
 | | The NMR structure of the MAX60 effector from Magnaporthe Oryzae | | 分子名称: | MAX effector protein | | 著者 | Lahfa, M, Padilla, A, de Guillen, K, Pissarra, J, Raji, M, Cesari, S, Kroj, T, Gladieux, P, Roumestand, C, Barthe, P. | | 登録日 | 2022-04-12 | | 公開日 | 2023-04-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | 1 H, 13 C, 15 N backbone and side-chain NMR assignments for three MAX effectors from Magnaporthe oryzae.
Biomol.Nmr Assign., 16, 2022
|
|
4MHV
 
 | | Crystal structure of the PNT domain of human ETS2 | | 分子名称: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | 著者 | Newman, J.A, Cooper, C.D.O, Krojer, T, Shrestha, L, Burgess-brown, N, Arrowsmith, C.H, Bountra, C, Edwards, A, Gileadi, O. | | 登録日 | 2013-08-30 | | 公開日 | 2013-09-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Crystal structure of the PNT domain of human ETS2
To be Published
|
|
4DIQ
 
 | | Crystal Structure of human NO66 | | 分子名称: | Lysine-specific demethylase NO66, NICKEL (II) ION, PYRIDINE-2,4-DICARBOXYLIC ACID, ... | | 著者 | Vollmar, M, Krojer, T, Ng, S, Pilka, E, Bray, J, Pike, A.C.W, Filippakopoulos, P, Roos, A, Arrowsmith, C.H, Edwards, E, Bountra, C, von Delft, F, Oppermann, U, Structural Genomics Consortium (SGC) | | 登録日 | 2012-01-31 | | 公開日 | 2012-03-07 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structure of human NO66
TO BE PUBLISHED
|
|
4UY4
 
 | | 1.86 A structure of human Spindlin-4 protein in complex with histone H3K4me3 peptide | | 分子名称: | GLYCEROL, HISTONE H3K4ME3, SPINDLIN-4 | | 著者 | Talon, R, Gileadi, C, Johansson, C, Burgess-Brown, N, Shrestha, L, von Delft, F, Krojer, T, Fairhead, M, Bountra, C, Arrowsmith, C.H, Edwards, A, Oppermann, U. | | 登録日 | 2014-08-28 | | 公開日 | 2014-09-24 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.862 Å) | | 主引用文献 | 1.86 A Structure of Human Spindlin-4 Protein in Complex with Histone H3K4Me3 Peptide
To be Published
|
|
4UUC
 
 | | Crystal structure of human ASB11 ankyrin repeat domain | | 分子名称: | ANKYRIN REPEAT AND SOCS BOX PROTEIN 11 | | 著者 | Pinkas, D.M, Sanvitale, C, Kragh Nielsen, T, Guo, K, Sorrell, F, Berridge, G, Ayinampudi, V, Wang, D, Newman, J.A, Tallant, C, Chaikuad, A, Canning, P, Kopec, J, Krojer, T, Vollmar, M, Allerston, C.K, Chalk, R, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Bullock, A. | | 登録日 | 2014-07-25 | | 公開日 | 2014-08-06 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of Human Asb11 Ankyrin Repeat Domain
To be Published
|
|
7B6K
 
 | | Crystal structure of MurE from E.coli in complex with Z57715447 | | 分子名称: | 5-cyclohexyl-3-(pyridin-4-yl)-1,2,4-oxadiazole, CITRIC ACID, DIMETHYL SULFOXIDE, ... | | 著者 | Koekemoer, L, Steindel, M, Fairhead, M, Talon, R, Douangamath, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | 登録日 | 2020-12-07 | | 公開日 | 2020-12-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.838 Å) | | 主引用文献 | Crystal structure of MurE from E.coli
To Be Published
|
|
