2YVA
 
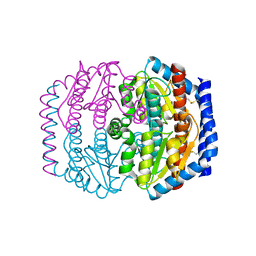 | | Crystal structure of Escherichia coli DiaA | | 分子名称: | DnaA initiator-associating protein diaA | | 著者 | Keyamura, K, Fujikawa, N, Ishida, T, Ozaki, S, Suetsugu, M, Kagawa, W, Yokoyama, S, Kurumizaka, H, Katayama, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-04-10 | | 公開日 | 2008-01-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The interaction of DiaA and DnaA regulates the replication cycle in E. coli by directly promoting ATP DnaA-specific initiation complexes
Genes Dev., 21, 2007
|
|
4XSH
 
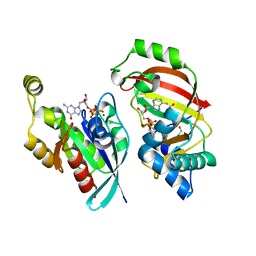 | | The complex structure of C3cer exoenzyme and GTP bound RhoA (NADH-bound state) | | 分子名称: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ... | | 著者 | Toda, A, Tsurumura, T, Yoshida, T, Tsuge, H. | | 登録日 | 2015-01-22 | | 公開日 | 2015-06-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Rho GTPase Recognition by C3 Exoenzyme Based on C3-RhoA Complex Structure.
J.Biol.Chem., 290, 2015
|
|
4XSG
 
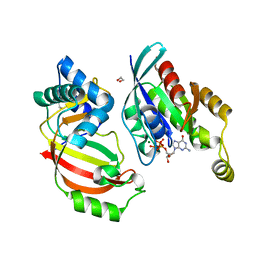 | | The complex structure of C3cer exoenzyme and GTP bound RhoA (NADH-free state) | | 分子名称: | 1,2-ETHANEDIOL, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ADP-ribosyltransferase, ... | | 著者 | Toda, A, Tsurumura, T, Yoshida, T, Tsuge, H. | | 登録日 | 2015-01-22 | | 公開日 | 2015-06-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Rho GTPase Recognition by C3 Exoenzyme Based on C3-RhoA Complex Structure.
J.Biol.Chem., 290, 2015
|
|
4ZM7
 
 | |
5AX6
 
 | | The crystal structure of CofB, the minor pilin subunit of CFA/III from human enterotoxigenic Escherichia coli. | | 分子名称: | ACETATE ION, CofB | | 著者 | Kawahara, K, Oki, K, Fukaksua, F, Maruno, T, Kobayashi, Y, Daisuke, M, Taniguchi, T, Honda, T, Iida, T, Nakamura, S, Ohkubo, T. | | 登録日 | 2015-07-16 | | 公開日 | 2016-03-09 | | 最終更新日 | 2020-02-26 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Homo-trimeric Structure of the Type IVb Minor Pilin CofB Suggests Mechanism of CFA/III Pilus Assembly in Human Enterotoxigenic Escherichia coli
J.Mol.Biol., 428, 2016
|
|
1JWQ
 
 | | Structure of the catalytic domain of CwlV, N-acetylmuramoyl-L-alanine amidase from Bacillus(Paenibacillus) polymyxa var.colistinus | | 分子名称: | N-ACETYLMURAMOYL-L-ALANINE AMIDASE CwlV, ZINC ION | | 著者 | Yamane, T, Koyama, Y, Nojiri, Y, Hikage, T, Akita, M, Suzuki, A, Shirai, T, Ise, F, Shida, T, Sekiguchi, J. | | 登録日 | 2001-09-05 | | 公開日 | 2003-11-18 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The Structure of the catalytic domain of N-acetylmuramoyl-L-alanine amidase, a cell wall hydrolase from Bacillus polymyxa var.colistinus and its resemblance to the structure of carboxypeptidases
To be Published
|
|
1MPY
 
 | | STRUCTURE OF CATECHOL 2,3-DIOXYGENASE (METAPYROCATECHASE) FROM PSEUDOMONAS PUTIDA MT-2 | | 分子名称: | ACETONE, CATECHOL 2,3-DIOXYGENASE, FE (II) ION | | 著者 | Kita, A, Kita, S, Fujisawa, I, Inaka, K, Ishida, T, Horiike, K, Nozaki, M, Miki, K. | | 登録日 | 1998-10-20 | | 公開日 | 1999-05-18 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | An archetypical extradiol-cleaving catecholic dioxygenase: the crystal structure of catechol 2,3-dioxygenase (metapyrocatechase) from Ppseudomonas putida mt-2.
Structure Fold.Des., 7, 1999
|
|
1MPT
 
 | | CRYSTAL STRUCTURE OF A NEW ALKALINE SERINE PROTEASE (M-PROTEASE) FROM BACILLUS SP. KSM-K16 | | 分子名称: | CALCIUM ION, M-PROTEASE | | 著者 | Yamane, T, Kani, T, Hatanaka, T, Suzuki, A, Ashida, T, Kobayashi, T, Ito, S, Yamashita, O. | | 登録日 | 1994-04-13 | | 公開日 | 1994-06-22 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure of a new alkaline serine protease (M-protease) from Bacillus sp. KSM-K16.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
5D45
 
 | | Crystal Structure of FABP4 in complex with 3-(5-cyclopropyl-2,3-diphenyl-1H-indol-1-yl)propanoic acid | | 分子名称: | 3-(5-cyclopropyl-2,3-diphenyl-1H-indol-1-yl)propanoic acid, Fatty acid-binding protein, adipocyte | | 著者 | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | 登録日 | 2015-08-07 | | 公開日 | 2016-06-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
5D4A
 
 | | Crystal Structure of FABP4 in complex with 3-(2-phenyl-1H-indol-1-yl)propanoic acid | | 分子名称: | 3-(2-phenyl-1H-indol-1-yl)propanoic acid, Fatty acid-binding protein, adipocyte | | 著者 | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | 登録日 | 2015-08-07 | | 公開日 | 2016-06-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
3MFP
 
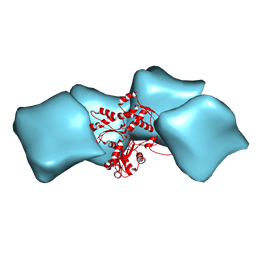 | | Atomic model of F-actin based on a 6.6 angstrom resolution cryoEM map | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle | | 著者 | Fujii, T, Iwane, A.H, Yanagida, T, Namba, K. | | 登録日 | 2010-04-03 | | 公開日 | 2010-09-29 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (6.6 Å) | | 主引用文献 | Direct visualization of secondary structures of F-actin by electron cryomicroscopy
Nature, 467, 2010
|
|
5D48
 
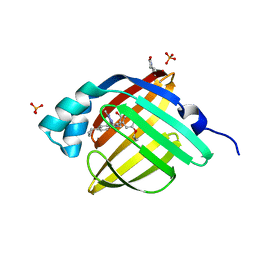 | | Crystal Structure of FABP4 in complex with 3-{5-cyclopropyl-3-(3,5-dimethyl-1H-pyrazol-4-yl)-2-[3-(propan-2-yloxy) phenyl]-1H-indol-1-yl}propanoic acid | | 分子名称: | 3-{5-cyclopropyl-3-(3,5-dimethyl-1H-pyrazol-4-yl)-2-[3-(propan-2-yloxy)phenyl]-1H-indol-1-yl}propanoic acid, Fatty acid-binding protein, adipocyte, ... | | 著者 | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | 登録日 | 2015-08-07 | | 公開日 | 2016-06-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
5D47
 
 | | Crystal Structure of FABP4 in complex with 3-[5-cyclopropyl-3-(3-methoxypyridin-4-yl)-2-phenyl-1H-indol-1-yl] propanoic acid | | 分子名称: | 3-[5-cyclopropyl-3-(3-methoxypyridin-4-yl)-2-phenyl-1H-indol-1-yl]propanoic acid, Fatty acid-binding protein, adipocyte | | 著者 | Tagami, U, Takahashi, K, Igarashi, S, Ejima, C, Yoshida, T, Takeshita, S, Miyanaga, W, Sugiki, M, Tokumasu, M, Hatanaka, T, Kashiwagi, T, Ishikawa, K, Miyano, H, Mizukoshi, T. | | 登録日 | 2015-08-07 | | 公開日 | 2016-06-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Interaction Analysis of FABP4 Inhibitors by X-ray Crystallography and Fragment Molecular Orbital Analysis
Acs Med.Chem.Lett., 7, 2016
|
|
5EXV
 
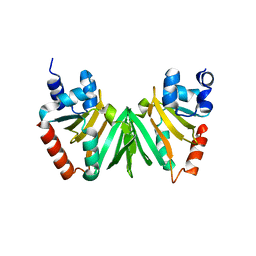 | |
5YQ0
 
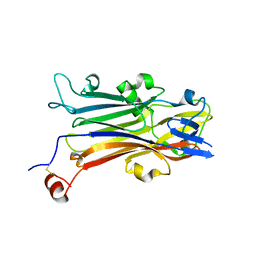 | | Crystal structure of secreted protein CofJ from ETEC. | | 分子名称: | CALCIUM ION, CofJ | | 著者 | Oki, H, Kawahara, K, Maruno, T, Imai, T, Muroga, Y, Fukakusa, S, Iwashita, T, Kobayashi, Y, Matsuda, S, Kodama, T, Iida, T, Yoshida, T, Ohkubo, T, Nakamura, S. | | 登録日 | 2017-11-04 | | 公開日 | 2018-06-27 | | 最終更新日 | 2018-07-25 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Interplay of a secreted protein with type IVb pilus for efficient enterotoxigenicEscherichia colicolonization
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YPZ
 
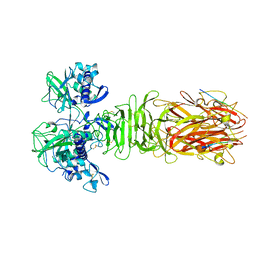 | | Crystal structure of minor pilin CofB from CFA/III complexed with N-terminal peptide fragment of CofJ | | 分子名称: | CofB, CofJ | | 著者 | Oki, H, Kawahara, K, Maruno, T, Imai, T, Muroga, Y, Fukakusa, S, Iwashita, T, Kobayashi, Y, Matsuda, S, Kodama, T, Iida, T, Yoshida, T, Ohkubo, T, Nakamura, S. | | 登録日 | 2017-11-04 | | 公開日 | 2018-06-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.521 Å) | | 主引用文献 | Interplay of a secreted protein with type IVb pilus for efficient enterotoxigenicEscherichia colicolonization.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5GTT
 
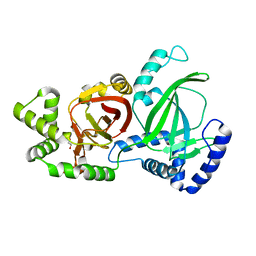 | | Crystal structure of C. perfringens iota-like enterotoxin CPILE-a | | 分子名称: | 1,2-ETHANEDIOL, Binary enterotoxin of Clostridium perfringens component a | | 著者 | Toniti, W, Yoshida, T, Tsurumura, T, Irikura, D, Tsuge, H. | | 登録日 | 2016-08-23 | | 公開日 | 2017-03-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.011 Å) | | 主引用文献 | Crystal structure and structure-based mutagenesis of actin-specific ADP-ribosylating toxin CPILE-a as novel enterotoxin
PLoS ONE, 12, 2017
|
|
5H6O
 
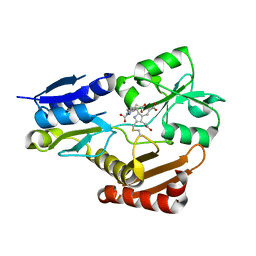 | | Porphobilinogen deaminase from Vibrio Cholerae | | 分子名称: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, MAGNESIUM ION, Porphobilinogen deaminase | | 著者 | Funamizu, T, Chen, M, Tanaka, Y, Ishimori, K, Uchida, T. | | 登録日 | 2016-11-14 | | 公開日 | 2017-11-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.702 Å) | | 主引用文献 | Porphobilinogen deaminase from Vibrio Cholerae
To Be Published
|
|
7VNN
 
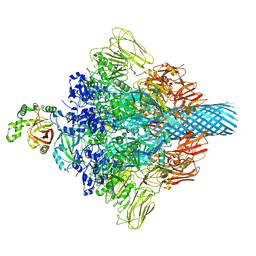 | | Complex structure of Clostridioides difficile enzymatic component (CDTa) and binding component (CDTb) pore with long stem | | 分子名称: | ADP-ribosylating binary toxin binding subunit CdtB, CALCIUM ION, CdtA | | 著者 | Yamada, T, Kawamoto, A, Yoshida, T, Sato, Y, Kato, T, Tsuge, H. | | 登録日 | 2021-10-11 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.64 Å) | | 主引用文献 | Cryo-EM structures of the translocational binary toxin complex CDTa-bound CDTb-pore from Clostridioides difficile.
Nat Commun, 13, 2022
|
|
7VNJ
 
 | | Complex structure of Clostridioides difficile enzymatic component (CDTa) and binding component (CDTb) pore with short stem | | 分子名称: | ADP-ribosylating binary toxin binding subunit CdtB, ADP-ribosyltransferase enzymatic component, CALCIUM ION | | 著者 | Yamada, T, Kawamoto, A, Yoshida, T, Sato, Y, Kato, T, Tsuge, H. | | 登録日 | 2021-10-11 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.56 Å) | | 主引用文献 | Cryo-EM structures of the translocational binary toxin complex CDTa-bound CDTb-pore from Clostridioides difficile.
Nat Commun, 13, 2022
|
|
2PA2
 
 | | Crystal structure of human Ribosomal protein L10 core domain | | 分子名称: | 60S ribosomal protein L10, POTASSIUM ION | | 著者 | Nishimura, M, Kaminishi, T, Takemoto, C, Kawazoe, M, Yoshida, T, Tanaka, A, Sugano, S, Shirouzu, M, Ohkubo, T, Yokoyama, S, Kobayashi, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-03-27 | | 公開日 | 2008-03-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of Human Ribosomal Protein L10 Core Domain Reveals Eukaryote-Specific Motifs in Addition to the Conserved Fold
J.Mol.Biol., 377, 2008
|
|
5BWM
 
 | | The complex structure of C3cer exoenzyme and GDP bound RhoA (NADH-bound state) | | 分子名称: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ADP-ribosyltransferase, ... | | 著者 | Toda, A, Tsurumura, T, Yoshida, T, Tsuge, H. | | 登録日 | 2015-06-08 | | 公開日 | 2015-06-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Rho GTPase Recognition by C3 Exoenzyme Based on C3-RhoA Complex Structure.
J.Biol.Chem., 290, 2015
|
|
7YVQ
 
 | | Complex structure of Clostridioides difficile binary toxin folded CDTa-bound CDTb-pore (short). | | 分子名称: | ADP-ribosylating binary toxin binding subunit CdtB, ADP-ribosylating binary toxin enzymatic subunit CdtA, CALCIUM ION | | 著者 | Yamada, T, Kawamoto, A, Yoshida, T, Sato, Y, Kato, T, Tsuge, H. | | 登録日 | 2022-08-19 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Cryo-EM structures of the translocational binary toxin complex CDTa-bound CDTb-pore from Clostridioides difficile.
Nat Commun, 13, 2022
|
|
7YVS
 
 | | Complex structure of Clostridioides difficile binary toxin unfolded CDTa-bound CDTb-pore (short). | | 分子名称: | ADP-ribosylating binary toxin binding subunit CdtB, ADP-ribosylating binary toxin enzymatic subunit CdtA, CALCIUM ION | | 著者 | Yamada, T, Kawamoto, A, Yoshida, T, Sato, Y, Kato, T, Tsuge, H. | | 登録日 | 2022-08-19 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Cryo-EM structures of the translocational binary toxin complex CDTa-bound CDTb-pore from Clostridioides difficile.
Nat Commun, 13, 2022
|
|
1KKT
 
 | | Structure of P. citrinum alpha 1,2-mannosidase reveals the basis for differences in specificity of the ER and Golgi Class I enzymes | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Mannosyl-oligosaccharide alpha-1,2-mannosidase, ... | | 著者 | Lobsanov, Y.D, Vallee, F, Imberty, A, Yoshida, T, Yip, P, Herscovics, A, Howell, P.L. | | 登録日 | 2001-12-10 | | 公開日 | 2002-01-23 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of Penicillium citrinum alpha 1,2-mannosidase reveals the basis for differences in specificity of the endoplasmic reticulum and Golgi class I enzymes.
J.Biol.Chem., 277, 2002
|
|
