3SUH
 
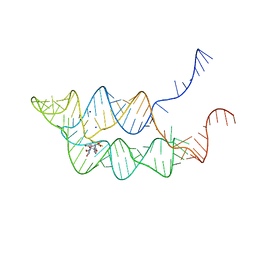 | | Crystal structure of THF riboswitch, bound with 5-formyl-THF | | 分子名称: | N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, Riboswitch, SODIUM ION | | 著者 | Huang, L, Serganov, A, Patel, D.J. | | 登録日 | 2011-07-11 | | 公開日 | 2011-09-14 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Long-range pseudoknot interactions dictate the regulatory response in the tetrahydrofolate riboswitch.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OXE
 
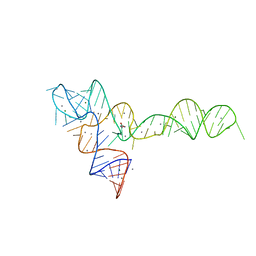 | | crystal structure of glycine riboswitch, Mn2+ soaked | | 分子名称: | GLYCINE, MAGNESIUM ION, MANGANESE (II) ION, ... | | 著者 | Huang, L, Serganov, A, Patel, D.J. | | 登録日 | 2010-09-21 | | 公開日 | 2010-12-29 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.899 Å) | | 主引用文献 | Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch.
Mol.Cell, 40, 2010
|
|
3OWZ
 
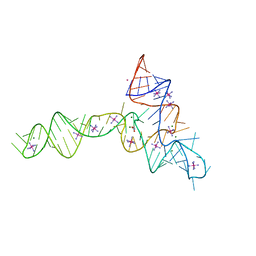 | | Crystal structure of glycine riboswitch, soaked in Iridium | | 分子名称: | Domain II of glycine riboswitch, GLYCINE, IRIDIUM HEXAMMINE ION, ... | | 著者 | Huang, L, Serganov, A, Patel, D.J. | | 登録日 | 2010-09-20 | | 公開日 | 2010-12-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.949 Å) | | 主引用文献 | Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch.
Mol.Cell, 40, 2010
|
|
3OXB
 
 | |
3OXM
 
 | | crystal structure of glycine riboswitch, Tl-Acetate soaked | | 分子名称: | GLYCINE, MAGNESIUM ION, THALLIUM (I) ION, ... | | 著者 | Huang, L, Serganov, A, Patel, D.J. | | 登録日 | 2010-09-21 | | 公開日 | 2010-12-29 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch.
Mol.Cell, 40, 2010
|
|
3OXJ
 
 | | crystal structure of glycine riboswitch, soaked in Ba2+ | | 分子名称: | BARIUM ION, GLYCINE, MAGNESIUM ION, ... | | 著者 | Huang, L, Serganov, A, Patel, D.J. | | 登録日 | 2010-09-21 | | 公開日 | 2010-12-29 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch.
Mol.Cell, 40, 2010
|
|
3OX0
 
 | |
3OWI
 
 | |
3OXD
 
 | |
3OWW
 
 | |
6HBT
 
 | |
6HBX
 
 | |
6HCT
 
 | |
6HC5
 
 | |
7XY1
 
 | |
7XYC
 
 | |
7Y3T
 
 | |
7Y22
 
 | |
7Y23
 
 | |
7Y1C
 
 | |
7Y5S
 
 | |
7EAG
 
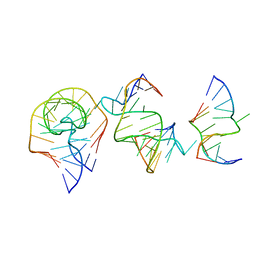 | | Crystal structure of the RAGATH-18 k-turn | | 分子名称: | RNA (5'-R(*GP*UP*CP*UP*AP*UP*GP*AP*AP*GP*GP*CP*UP*GP*GP*AP*GP*AP*C)-3') | | 著者 | Huang, L, Lilley, D.M.J. | | 登録日 | 2021-03-07 | | 公開日 | 2021-06-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure and folding of four putative kink turns identified in structured RNA species in a test of structural prediction rules.
Nucleic Acids Res., 49, 2021
|
|
7EAF
 
 | |
6L62
 
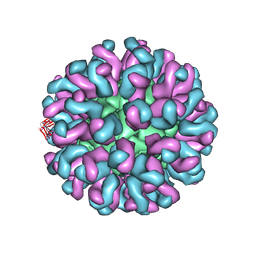 | | Neutralization mechanism of a monoclonal antibody targeting a porcine circovirus type 2 Cap protein conformational epitope | | 分子名称: | Capsid protein, Heavy chain of Fab fragment, Light chain of Fab fragment | | 著者 | Sun, Z, Huang, L, Xia, D, Wei, Y, Sun, E, Zhu, H, Bian, H, Wu, H, Feng, L, Wang, J, Liu, C. | | 登録日 | 2019-10-25 | | 公開日 | 2020-02-12 | | 最終更新日 | 2020-04-29 | | 実験手法 | ELECTRON MICROSCOPY (7.2 Å) | | 主引用文献 | Neutralization Mechanism of a Monoclonal Antibody Targeting a Porcine Circovirus Type 2 Cap Protein Conformational Epitope.
J.Virol., 94, 2020
|
|
6Q8U
 
 | |
