5AUS
 
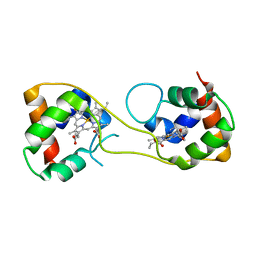 | | Hydrogenobacter thermophilus cytochrome c552 dimer formed by domain swapping at C-terminal region | | 分子名称: | Cytochrome c-552, HEME C | | 著者 | Ren, C, Nagao, S, Yamanaka, M, Komori, H, Shomura, Y, Higuchi, Y, Hirota, S. | | 登録日 | 2015-06-08 | | 公開日 | 2015-10-21 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Oligomerization enhancement and two domain swapping mode detection for thermostable cytochrome c552via the elongation of the major hinge loop.
Mol Biosyst, 11, 2015
|
|
5GYR
 
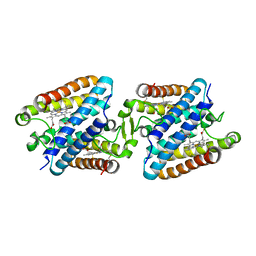 | | Tetrameric Allochromatium vinosum cytochrome c' | | 分子名称: | Cytochrome c', HEME C | | 著者 | Yamanaka, M, Hoshizumi, M, Nagao, S, Nakayama, R, Shibata, N, Higuchi, Y, Hirota, S. | | 登録日 | 2016-09-23 | | 公開日 | 2017-02-08 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Formation and carbon monoxide-dependent dissociation of Allochromatium vinosum cytochrome c' oligomers using domain-swapped dimers
Protein Sci., 26, 2017
|
|
1A7V
 
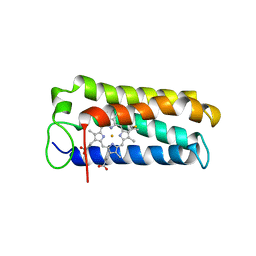 | | CYTOCHROME C' FROM RHODOPSEUDOMONAS PALUSTRIS | | 分子名称: | CYTOCHROME C', PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Shibata, N, Iba, S, Misaki, S, Meyer, T.E, Bartsch, R.G, Cusanovich, M.A, Higuchi, Y, Yasuoka, N. | | 登録日 | 1998-03-18 | | 公開日 | 1998-06-17 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Basis for monomer stabilization in Rhodopseudomonas palustris cytochrome c' derived from the crystal structure.
J.Mol.Biol., 284, 1998
|
|
4E1O
 
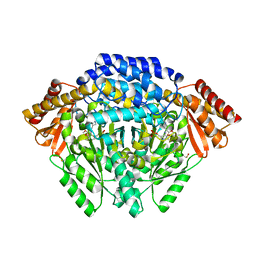 | | Human histidine decarboxylase complex with Histidine methyl ester (HME) | | 分子名称: | HISTIDINE-METHYL-ESTER, Histidine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Komori, H, Nitta, Y, Ueno, H, Higuchi, Y. | | 登録日 | 2012-03-06 | | 公開日 | 2012-07-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural study reveals that Ser-354 determines substrate specificity on human histidine decarboxylase
J.Biol.Chem., 287, 2012
|
|
1WQ2
 
 | | Neutron Crystal Structure Of Dissimilatory Sulfite Reductase D (DsrD) | | 分子名称: | Protein dsvD, SULFATE ION | | 著者 | Chatake, T, Mizuno, N, Voordouw, G, Higuchi, Y, Arai, S, Tanaka, I, Niimura, N. | | 登録日 | 2004-09-19 | | 公開日 | 2005-09-19 | | 最終更新日 | 2023-10-25 | | 実験手法 | NEUTRON DIFFRACTION (2.4 Å) | | 主引用文献 | Crystallization and preliminary neutron analysis of the dissimilatory sulfite reductase D (DsrD) protein from the sulfate-reducing bacterium Desulfovibrio vulgaris.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
7VXQ
 
 | | The Carbon Monoxide Complex of [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 | | 分子名称: | CARBON MONOXIDE, FE3-S4 CLUSTER, GLYCEROL, ... | | 著者 | Nishikawa, K, Higuchi, K, Imanishi, T, Higuchi, Y. | | 登録日 | 2021-11-13 | | 公開日 | 2022-02-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Structural and spectroscopic characterization of CO inhibition of [NiFe]-hydrogenase from Citrobacter sp. S-77.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
3P1P
 
 | | Crystal structure of human 14-3-3 sigma C38N/N166H in complex with TASK-3 peptide | | 分子名称: | 14-3-3 protein sigma, 6-mer peptide from Potassium channel subfamily K member 9, CHLORIDE ION, ... | | 著者 | Anders, C, Higuchi, Y, Schumacher, B, Thiel, P, Kato, N, Ottmann, C. | | 登録日 | 2010-09-30 | | 公開日 | 2011-10-12 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | A semisynthetic fusicoccane stabilizes a protein-protein interaction and enhances the expression of K+ channels at the cell surface
Chem. Biol., 20, 2013
|
|
3P1Q
 
 | | Crystal structure of human 14-3-3 sigma C38N/N166H in complex with TASK-3 Peptide and stabilizer fusicoccin A | | 分子名称: | 14-3-3 protein sigma, 6-mer peptide from Potassium channel subfamily K member 9, CALCIUM ION, ... | | 著者 | Anders, C, Higuchi, Y, Schumacher, B, Thiel, P, Kato, N, Ottmann, C. | | 登録日 | 2010-09-30 | | 公開日 | 2011-10-12 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | A semisynthetic fusicoccane stabilizes a protein-protein interaction and enhances the expression of K+ channels at the cell surface
Chem. Biol., 20, 2013
|
|
3P1O
 
 | | Crystal structure of human 14-3-3 sigma in complex with TASK-3 peptide and stabilisator Fusicoccin A | | 分子名称: | 14-3-3 protein sigma, 6-mer peptide from Potassium channel subfamily K member 9, CALCIUM ION, ... | | 著者 | Anders, C, Higuchi, Y, Schumacher, B, Thiel, P, Kato, N, Ottmann, C. | | 登録日 | 2010-09-30 | | 公開日 | 2011-10-12 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A semisynthetic fusicoccane stabilizes a protein-protein interaction and enhances the expression of K+ channels at the cell surface
Chem. Biol., 20, 2013
|
|
3P1N
 
 | | Crystal structure of human 14-3-3 sigma in complex with TASK-3 peptide | | 分子名称: | 14-3-3 protein sigma, 6-mer peptide from Potassium channel subfamily K member 9, CHLORIDE ION, ... | | 著者 | Anders, C, Higuchi, Y, Schumacher, B, Thiel, P, Kato, N, Ottmann, C. | | 登録日 | 2010-09-30 | | 公開日 | 2011-10-12 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | A semisynthetic fusicoccane stabilizes a protein-protein interaction and enhances the expression of K+ channels at the cell surface
Chem. Biol., 20, 2013
|
|
3P1S
 
 | | Crystal structure of human 14-3-3 sigma C38N/N166H in complex with TASK-3 peptide and stabilizer fusicoccin A | | 分子名称: | 14-3-3 protein sigma, 6-mer peptide from Potassium channel subfamily K member 9, CHLORIDE ION, ... | | 著者 | Anders, C, Higuchi, Y, Schumacher, B, Thiel, P, Kato, N, Ottmann, C. | | 登録日 | 2010-09-30 | | 公開日 | 2011-10-12 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | A semisynthetic fusicoccane stabilizes a protein-protein interaction and enhances the expression of K+ channels at the cell surface
Chem. Biol., 20, 2013
|
|
3P1R
 
 | | Crystal structure of human 14-3-3 sigma C38V/N166H in complex with TASK-3 peptide | | 分子名称: | 14-3-3 protein sigma, 6-mer peptide from Potassium channel subfamily K member 9, CALCIUM ION, ... | | 著者 | Anders, C, Higuchi, Y, Schumacher, B, Thiel, P, Kato, N, Ottmann, C. | | 登録日 | 2010-09-30 | | 公開日 | 2011-10-12 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | A semisynthetic fusicoccane stabilizes a protein-protein interaction and enhances the expression of K+ channels at the cell surface
Chem. Biol., 20, 2013
|
|
3LZW
 
 | | Crystal structure of ferredoxin-NADP+ oxidoreductase from bacillus subtilis (form I) | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase 2, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Komori, H, Seo, D, Sakurai, T, Higuchi, Y. | | 登録日 | 2010-03-02 | | 公開日 | 2010-12-08 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure analysis of Bacillus subtilis ferredoxin-NADP(+) oxidoreductase and the structural basis for its substrate selectivity
Protein Sci., 19, 2010
|
|
5B7F
 
 | | Structure of CueO - the signal peptide was truncated by HRV3C protease | | 分子名称: | 1,2-ETHANEDIOL, Blue copper oxidase CueO, CALCIUM ION, ... | | 著者 | Akter, M, Higuchi, Y, Shibata, N. | | 登録日 | 2016-06-07 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Biochemical, spectroscopic and X-ray structural analysis of deuterated multicopper oxidase CueO prepared from a new expression construct for neutron crystallography
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5B7M
 
 | | Structure of perdeuterated CueO - the signal peptide was truncated by HRV3C protease | | 分子名称: | Blue copper oxidase CueO, COPPER (II) ION | | 著者 | Akter, M, Higuchi, Y, Shibata, N. | | 登録日 | 2016-06-07 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Biochemical, spectroscopic and X-ray structural analysis of deuterated multicopper oxidase CueO prepared from a new expression construct for neutron crystallography
Acta Crystallogr.,Sect.F, 72, 2016
|
|
1WR5
 
 | |
7YH6
 
 | | Structure of SARS-CoV-2 spike RBD in complex with neutralizing antibody NIV-8 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-8 Fab heavy chain, NIV-8 Fab light chain, ... | | 著者 | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | 登録日 | 2022-07-12 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
7YH7
 
 | | SARS-CoV-2 spike in complex with neutralizing antibody NIV-8 (state 2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-8 Fab heavy chain, ... | | 著者 | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | 登録日 | 2022-07-13 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
5B7E
 
 | | Structure of perdeuterated CueO | | 分子名称: | Blue copper oxidase CueO, COPPER (II) ION, CU-O-CU LINKAGE, ... | | 著者 | Akter, M, Higuchi, Y, Shibata, N. | | 登録日 | 2016-06-07 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Biochemical, spectroscopic and X-ray structural analysis of deuterated multicopper oxidase CueO prepared from a new expression construct for neutron crystallography
Acta Crystallogr.,Sect.F, 72, 2016
|
|
3VWQ
 
 | | 6-aminohexanoate-dimer hydrolase S112A/G181D/R187A/H266N/D370Y mutant complexd with 6-aminohexanoate | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | 著者 | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | 登録日 | 2012-08-30 | | 公開日 | 2013-10-16 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
to be published
|
|
3VWP
 
 | | Crystal structure of 6-aminohexanoate-dimer hydrolase S112A/G181D/R187S/H266N/D370Y mutant complexd with 6-aminohexanoate | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | 著者 | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | 登録日 | 2012-08-30 | | 公開日 | 2013-10-16 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
to be published
|
|
1UBH
 
 | | Three-dimensional Structure of The Carbon Monoxide Complex of [NiFe]hydrogenase From Desulufovibrio vulgaris Miyazaki F | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, (MU-SULPHIDO)-BIS(MU-CYS,S)-[TRICARBONYLIRON-DI-(CYS,S)NICKEL(II)](FE-NI), CARBON MONOXIDE, ... | | 著者 | Ogata, H, Mizoguchi, Y, Mizuno, N, Miki, K, Adachi, S, Yasuoka, N, Yagi, T, Yamauchi, O, Hirota, S, Higuchi, Y. | | 登録日 | 2003-04-04 | | 公開日 | 2003-04-29 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Structural Studies of the Carbon Monoxide Complex of [NiFe]hydrogenase from Desulfovibrio vulgaris Miyazaki F: Suggestion for the Initial Activation Site for Dihydrogen
J.Am.Chem.Soc., 124, 2002
|
|
4E9Y
 
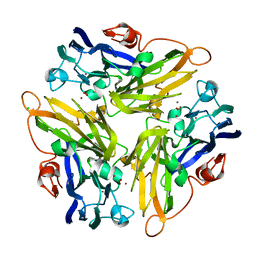 | | Multicopper Oxidase mgLAC (data4) | | 分子名称: | CHLORIDE ION, COPPER (II) ION, Multicopper oxidase, ... | | 著者 | Komori, H, Miyazaki, K, Higuchi, Y. | | 登録日 | 2012-03-21 | | 公開日 | 2013-03-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | New insights into the catalytic active-site structure of multicopper oxidases.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3VWN
 
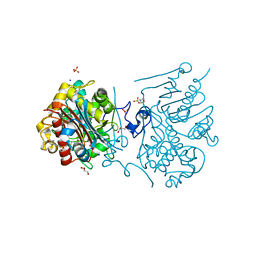 | | Crystal structure of 6-aminohexanoate-dimer hydrolase G181D/R187G/H266N/D370Y mutant | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | 著者 | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | 登録日 | 2012-08-30 | | 公開日 | 2013-10-16 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
To be Published
|
|
4E9W
 
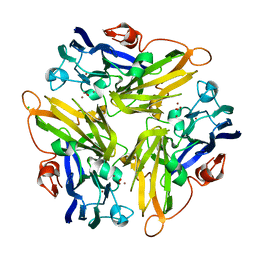 | | Multicopper Oxidase mgLAC (data2) | | 分子名称: | CHLORIDE ION, COPPER (II) ION, Multicopper oxidase, ... | | 著者 | Komori, H, Miyazaki, K, Higuchi, Y. | | 登録日 | 2012-03-21 | | 公開日 | 2013-03-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | New insights into the catalytic active-site structure of multicopper oxidases.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
