5H63
 
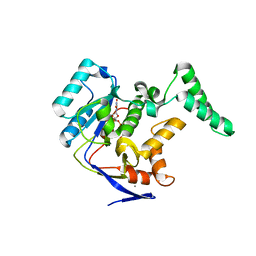 | | Structure of Transferase mutant-C23S,C199S | | 分子名称: | MANGANESE (II) ION, Transferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | 著者 | Park, J.B, Yoo, Y, Kim, J. | | 登録日 | 2016-11-10 | | 公開日 | 2017-12-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Structural basis for arginine glycosylation of host substrates by bacterial effector proteins.
Nat Commun, 9, 2018
|
|
5H62
 
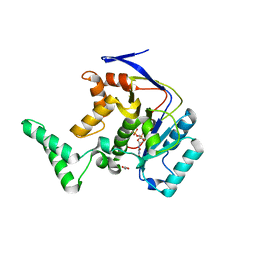 | | Structure of Transferase mutant-C23S,C199S | | 分子名称: | 1,2-ETHANEDIOL, MANGANESE (II) ION, Transferase, ... | | 著者 | Park, J.B, Yoo, Y, Kim, J. | | 登録日 | 2016-11-10 | | 公開日 | 2017-12-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Structural basis for arginine glycosylation of host substrates by bacterial effector proteins.
Nat Commun, 9, 2018
|
|
5H60
 
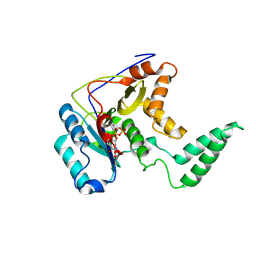 | | Structure of Transferase mutant-C23S,C199S | | 分子名称: | MANGANESE (II) ION, Transferase, URIDINE-5'-DIPHOSPHATE | | 著者 | Park, J.B, Yoo, Y, Kim, J. | | 登録日 | 2016-11-10 | | 公開日 | 2017-12-20 | | 最終更新日 | 2018-10-31 | | 実験手法 | X-RAY DIFFRACTION (3.64 Å) | | 主引用文献 | Structural basis for arginine glycosylation of host substrates by bacterial effector proteins.
Nat Commun, 9, 2018
|
|
5H61
 
 | |
5H5Y
 
 | |
6XJF
 
 | |
6PLN
 
 | |
6A6K
 
 | | Crystal structure of Estrogen-related Receptor-3 (ERR-gamma) ligand binding domain with DN201000 | | 分子名称: | 3-[(~{E})-5-oxidanyl-2-phenyl-1-[4-(4-propan-2-ylpiperazin-1-yl)phenyl]pent-1-enyl]phenol, Estrogen-related receptor gamma | | 著者 | Yoon, H, Kim, J, Chin, J, Cho, S.J, Song, J. | | 登録日 | 2018-06-28 | | 公開日 | 2019-04-10 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Discovery of Potent, Selective, and Orally Bioavailable Estrogen-Related Receptor-gamma Inverse Agonists To Restore the Sodium Iodide Symporter Function in Anaplastic Thyroid Cancer.
J. Med. Chem., 62, 2019
|
|
1DMQ
 
 | | CRYSTAL STRUCTURE OF MUTANT ENZYME Y32F OF KETOSTEROID ISOMERASE FROM PSEUDOMONAS PUTIDA BIOTYPE B | | 分子名称: | STEROID DELTA-ISOMERASE | | 著者 | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | 登録日 | 1999-12-14 | | 公開日 | 2000-05-23 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Contribution of the hydrogen-bond network involving a tyrosine triad in the active site to the structure and function of a highly proficient ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 39, 2000
|
|
1DMM
 
 | | CRYSTAL STRUCTURES OF MUTANT ENZYMES Y57F OF KETOSTEROID ISOMERASE FROM PSEUDOMONAS PUTIDA BIOTYPE B | | 分子名称: | STEROID DELTA-ISOMERASE | | 著者 | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | 登録日 | 1999-12-14 | | 公開日 | 2000-05-23 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Contribution of the hydrogen-bond network involving a tyrosine triad in the active site to the structure and function of a highly proficient ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 39, 2000
|
|
1DMN
 
 | | CRYSTAL STRUCTURE OF MUTANT ENZYME Y32F/Y57F OF KETOSTEROID ISOMERASE FROM PSEUDOMONAS PUTIDA BIOTYPE B | | 分子名称: | STEROID DELTA-ISOMERASE | | 著者 | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | 登録日 | 1999-12-14 | | 公開日 | 2000-05-23 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Contribution of the hydrogen-bond network involving a tyrosine triad in the active site to the structure and function of a highly proficient ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 39, 2000
|
|
5G54
 
 | | The crystal structure of light-driven chloride pump ClR at pH 4.5 | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | 登録日 | 2016-05-19 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an Ntq Motif.
Nat.Commun., 7, 2016
|
|
5G28
 
 | | The crystal structure of light-driven chloride pump ClR at pH 6.0. | | 分子名称: | CHLORIDE ION, CHLORIDE PUMPING RHODOPSIN, OLEIC ACID, ... | | 著者 | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | 登録日 | 2016-04-07 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an Ntq Motif.
Nat.Commun., 7, 2016
|
|
5G2D
 
 | | The crystal structure of light-driven chloride pump ClR (T102N) mutant at pH 4.5. | | 分子名称: | CHLORIDE ION, CHLORIDE PUMP RHODOPSIN, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | 登録日 | 2016-04-07 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an Ntq Motif.
Nat.Commun., 7, 2016
|
|
5G2A
 
 | | The crystal structure of light-driven chloride pump ClR at pH 6.0 with Bromide ion. | | 分子名称: | BROMIDE ION, CHLORIDE PUMPING RHODOPSIN, RETINAL | | 著者 | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | 登録日 | 2016-04-07 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an Ntq Motif.
Nat.Commun., 7, 2016
|
|
5G2C
 
 | | The crystal structure of light-driven chloride pump ClR (T102D) mutant at pH 4.5. | | 分子名称: | CHLORIDE ION, CHLORIDE PUMPING RHODOPSIN, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | 登録日 | 2016-04-07 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an Ntq Motif.
Nat.Commun., 7, 2016
|
|
5ZTK
 
 | | Synchrotron structure of light-driven chloride pump having an NTQ motif | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Park, J.H, Park, S.Y, Lee, W. | | 登録日 | 2018-05-04 | | 公開日 | 2018-12-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Non-cryogenic structure of a chloride pump provides crucial clues to temperature-dependent channel transport efficiency
J. Biol. Chem., 294, 2019
|
|
5ZTL
 
 | | Non-cryogenic structure of light-driven chloride pump having an NTQ motif | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Park, S.Y, Liu, H, Lee, W. | | 登録日 | 2018-05-04 | | 公開日 | 2018-12-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Non-cryogenic structure of a chloride pump provides crucial clues to temperature-dependent channel transport efficiency
J. Biol. Chem., 294, 2019
|
|
7EOY
 
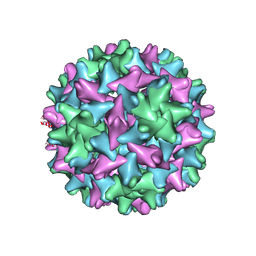 | | Engineered Hepatitis B virus core antigen T=3 | | 分子名称: | Capsid protein,Immunoglobulin G-binding protein A | | 著者 | Jeong, H, Heo, Y, Yoo, Y, Ryu, B, Yun, J, Cho, H, Lee, W. | | 登録日 | 2021-04-24 | | 公開日 | 2021-09-01 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural and Functional Characterizations of Cancer Targeting Nanoparticles Based on Hepatitis B Virus Capsid.
Int J Mol Sci, 22, 2021
|
|
7FDJ
 
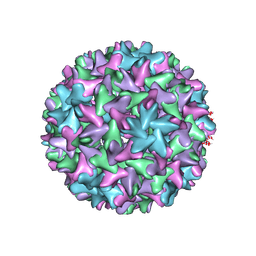 | | Engineered Hepatitis B virus core antigen with short linker T=4 | | 分子名称: | Capsid protein,Immunoglobulin G-binding protein A | | 著者 | Jeong, H, Heo, Y, Yoo, Y, Ryu, B, Yun, J, Cho, H, Lee, W. | | 登録日 | 2021-07-16 | | 公開日 | 2021-09-01 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Structural and Functional Characterizations of Cancer Targeting Nanoparticles Based on Hepatitis B Virus Capsid.
Int J Mol Sci, 22, 2021
|
|
7EP6
 
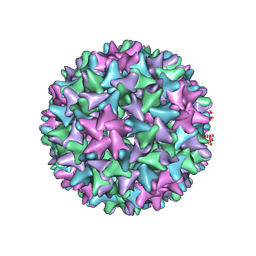 | | Engineered Hepatitis B virus core antigen T=4 | | 分子名称: | Capsid protein,Immunoglobulin G-binding protein A | | 著者 | Jeong, H, Heo, Y, Yoo, Y, Ryu, B, Yun, J, Cho, H, Lee, W. | | 登録日 | 2021-04-26 | | 公開日 | 2021-09-01 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.86 Å) | | 主引用文献 | Structural and Functional Characterizations of Cancer Targeting Nanoparticles Based on Hepatitis B Virus Capsid.
Int J Mol Sci, 22, 2021
|
|
1SMA
 
 | |
7VYR
 
 | |
1ZDM
 
 | | Crystal Structure of Activated CheY Bound to Xe | | 分子名称: | Chemotaxis protein cheY, MANGANESE (II) ION, XENON | | 著者 | Lowery, T.J, Doucleff, M, Ruiz, E.J, Rubin, S.M, Pines, A, Wemmer, D.E. | | 登録日 | 2005-04-14 | | 公開日 | 2005-04-26 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Distinguishing multiple chemotaxis Y protein conformations with laser-polarized 129Xe NMR.
Protein Sci., 14, 2005
|
|
6KF4
 
 | | Cryo-EM structure of Thermococcus kodakarensis RNA polymerase | | 分子名称: | DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit A'', DNA-directed RNA polymerase subunit D, ... | | 著者 | Jun, S.-H, Hyun, J, Jeong, H, Cha, J.S, Kim, H, Bartlett, M.S, Cho, H.-S, Murakami, K.S. | | 登録日 | 2019-07-06 | | 公開日 | 2020-07-01 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.97 Å) | | 主引用文献 | Direct binding of TFE alpha opens DNA binding cleft of RNA polymerase.
Nat Commun, 11, 2020
|
|
