1DMS
 
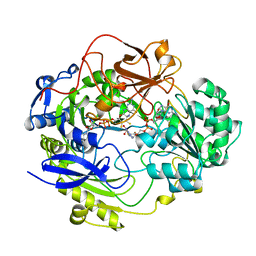 | | STRUCTURE OF DMSO REDUCTASE | | 分子名称: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DMSO REDUCTASE, MOLYBDENUM (IV)OXIDE | | 著者 | Schneider, F, Loewe, J, Huber, R, Schindelin, H, Kisker, C, Knaeblein, J. | | 登録日 | 1996-09-03 | | 公開日 | 1998-07-01 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Crystal structure of dimethyl sulfoxide reductase from Rhodobacter capsulatus at 1.88 A resolution.
J.Mol.Biol., 263, 1996
|
|
1QQ0
 
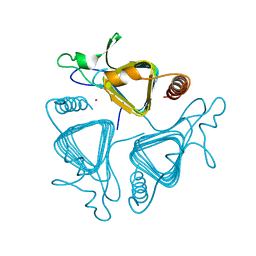 | | COBALT SUBSTITUTED CARBONIC ANHYDRASE FROM METHANOSARCINA THERMOPHILA | | 分子名称: | CARBONIC ANHYDRASE, COBALT (II) ION | | 著者 | Iverson, T.M, Alber, B.E, Kisker, C, Ferry, J.G, Rees, D.C. | | 登録日 | 1999-06-10 | | 公開日 | 1999-06-24 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | A closer look at the active site of gamma-class carbonic anhydrases: high-resolution crystallographic studies of the carbonic anhydrase from Methanosarcina thermophila.
Biochemistry, 39, 2000
|
|
5I7V
 
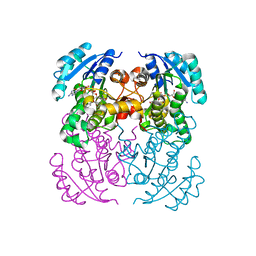 | | Crystal structure of B. pseudomallei FabI in complex with NAD and PT02 | | 分子名称: | 2-phenoxy-5-propyl-phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Hirschbeck, M.W, Eltschkner, S, Tonge, P.J, Kisker, C. | | 登録日 | 2016-02-18 | | 公開日 | 2017-02-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Rationalizing the Binding Kinetics for the Inhibition of the Burkholderia pseudomallei FabI1 Enoyl-ACP Reductase.
Biochemistry, 56, 2017
|
|
5I9L
 
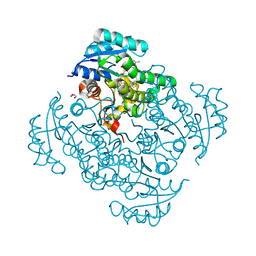 | | Crystal structure of B. pseudomallei FabI in complex with NAD and PT404 | | 分子名称: | 2-(2-chloro-4-nitrophenoxy)-5-ethyl-4-fluorophenol, Enoyl-[acyl-carrier-protein] reductase [NADH], GLYCEROL, ... | | 著者 | Hirschbeck, M.W, Eltschkner, S, Tonge, P.J, Kisker, C. | | 登録日 | 2016-02-20 | | 公開日 | 2017-02-22 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Rationalizing the Binding Kinetics for the Inhibition of the Burkholderia pseudomallei FabI1 Enoyl-ACP Reductase.
Biochemistry, 56, 2017
|
|
1D9X
 
 | | CRYSTAL STRUCTURE OF THE DNA REPAIR PROTEIN UVRB | | 分子名称: | EXCINUCLEASE UVRABC COMPONENT UVRB, ZINC ION | | 著者 | Theis, K, Chen, P.J, Skorvaga, M, Van Houten, B, Kisker, C. | | 登録日 | 1999-10-30 | | 公開日 | 2000-05-03 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of UvrB, a DNA helicase adapted for nucleotide excision repair.
EMBO J., 18, 1999
|
|
1QRE
 
 | | A CLOSER LOOK AT THE ACTIVE SITE OF GAMMA-CARBONIC ANHYDRASES: HIGH RESOLUTION CRYSTALLOGRAPHIC STUDIES OF THE CARBONIC ANHYDRASE FROM METHANOSARCINA THERMOPHILA | | 分子名称: | BICARBONATE ION, CARBONIC ANHYDRASE, COBALT (II) ION | | 著者 | Iverson, T.M, Alber, B.E, Kisker, C, Ferry, J.G, Rees, D.C. | | 登録日 | 1999-06-13 | | 公開日 | 1999-06-25 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | A closer look at the active site of gamma-class carbonic anhydrases: high-resolution crystallographic studies of the carbonic anhydrase from Methanosarcina thermophila.
Biochemistry, 39, 2000
|
|
1QRL
 
 | | A CLOSER LOOK AT THE ACTIVE SITE OF GAMMA-CARBONIC ANHYDRASES: HIGH RESOLUTION CRYSTALLOGRAPHIC STUDIES OF THE CARBONIC ANHYDRASE FROM METHANOSARCINA THERMOPHILA | | 分子名称: | BICARBONATE ION, CARBONIC ANHYDRASE, ZINC ION | | 著者 | Iverson, T.M, Alber, B.E, Kisker, C, Ferry, J.G, Rees, D.C. | | 登録日 | 1999-06-15 | | 公開日 | 1999-06-25 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | A closer look at the active site of gamma-class carbonic anhydrases: high-resolution crystallographic studies of the carbonic anhydrase from Methanosarcina thermophila.
Biochemistry, 39, 2000
|
|
1QRG
 
 | | A CLOSER LOOK AND THE ACTIVE SITE OF GAMMA-CARBONIC ANHYDRASES: HIGH RESOLUTION CRYSTALLOGRAPHIC STUDIES OF THE CARBONIC ANHYDRASE FROM METHANOSARCINA THERMOPHILA | | 分子名称: | CARBONIC ANHYDRASE, ZINC ION | | 著者 | Iverson, T.M, Alber, B.E, Kisker, C, Ferry, J.G, Rees, D.C. | | 登録日 | 1999-06-13 | | 公開日 | 1999-06-25 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | A closer look at the active site of gamma-class carbonic anhydrases: high-resolution crystallographic studies of the carbonic anhydrase from Methanosarcina thermophila.
Biochemistry, 39, 2000
|
|
1Q51
 
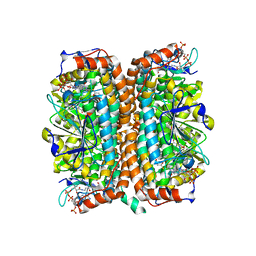 | | Crystal Structure of Mycobacterium tuberculosis MenB in Complex with Acetoacetyl-Coenzyme A, a Key Enzyme in Vitamin K2 Biosynthesis | | 分子名称: | ACETOACETYL-COENZYME A, menB | | 著者 | Truglio, J.J, Theis, K, Feng, Y, Gajda, R, Machutta, C, Tonge, P.J, Kisker, C, TB Structural Genomics Consortium (TBSGC) | | 登録日 | 2003-08-05 | | 公開日 | 2004-01-27 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of Mycobacterium tuberculosis MenB, a key enzyme in vitamin K2 biosynthesis.
J.Biol.Chem., 278, 2003
|
|
1D9Z
 
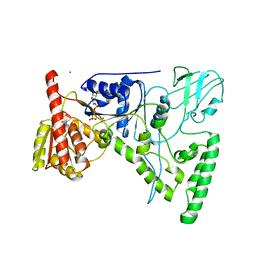 | | CRYSTAL STRUCTURE OF THE DNA REPAIR PROTEIN UVRB IN COMPLEX WITH ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, EXCINUCLEASE UVRABC COMPONENT UVRB, MAGNESIUM ION, ... | | 著者 | Theis, K, Chen, P.J, Skorvaga, M, Van Houten, B, Kisker, C. | | 登録日 | 1999-10-30 | | 公開日 | 2000-05-03 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Crystal structure of UvrB, a DNA helicase adapted for nucleotide excision repair.
EMBO J., 18, 1999
|
|
6ENS
 
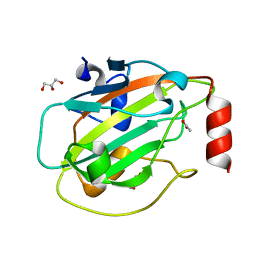 | | Structure of mouse wild-type RKIP | | 分子名称: | ACETATE ION, GLYCEROL, Phosphatidylethanolamine-binding protein 1 | | 著者 | Hirschbeck, M, Koelmel, W, Schindelin, H, Lorenz, K, Kisker, C. | | 登録日 | 2017-10-06 | | 公開日 | 2017-12-13 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Conserved salt-bridge competition triggered by phosphorylation regulates the protein interactome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5OBZ
 
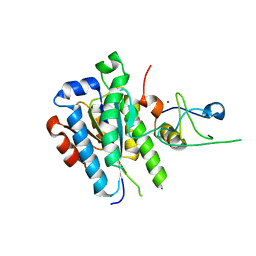 | | low resolution structure of the p34ct/p44ct minimal complex | | 分子名称: | Putative transcription factor, ZINC ION | | 著者 | Schoenwetter, E, Koelmel, W, Schmitt, D.R, Kuper, J, Kisker, C. | | 登録日 | 2017-06-29 | | 公開日 | 2017-10-18 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | The intricate network between the p34 and p44 subunits is central to the activity of the transcription/DNA repair factor TFIIH.
Nucleic Acids Res., 45, 2017
|
|
5NUS
 
 | | Structure of a minimal complex between p44 and p34 from Chaetomium thermophilum | | 分子名称: | ZINC ION, p34, p44 | | 著者 | Koelmel, W, Schoenwetter, E, Kuper, J, Schmitt, D.R, Kisker, C. | | 登録日 | 2017-05-02 | | 公開日 | 2017-10-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The intricate network between the p34 and p44 subunits is central to the activity of the transcription/DNA repair factor TFIIH.
Nucleic Acids Res., 45, 2017
|
|
2EUA
 
 | | Structure and Mechanism of MenF, the Menaquinone-Specific Isochorismate Synthase from Escherichia Coli | | 分子名称: | D(-)-TARTARIC ACID, Menaquinone-specific isochorismate synthase | | 著者 | Kolappan, S, Kisker, C, Zwahlen, J, Zhou, R, Tonge, P.J. | | 登録日 | 2005-10-28 | | 公開日 | 2006-12-05 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Lysine 190 is the catalytic base in MenF, the menaquinone-specific isochorismate synthase from Escherichia coli: implications for an enzyme family.
Biochemistry, 46, 2007
|
|
2FDC
 
 | |
2F4M
 
 | | The Mouse PNGase-HR23 Complex Reveals a Complete Remodulation of the Protein-Protein Interface Compared to its Yeast Orthologs | | 分子名称: | CHLORIDE ION, UV excision repair protein RAD23 homolog B, ZINC ION, ... | | 著者 | Zhao, G, Zhou, X, Wang, L, Kisker, C, Lennarz, W.J, Schindelin, H. | | 登録日 | 2005-11-23 | | 公開日 | 2006-03-07 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structure of the mouse peptide N-glycanase-HR23 complex suggests co-evolution of the endoplasmic reticulum-associated degradation and DNA repair pathways.
J.Biol.Chem., 281, 2006
|
|
2F4O
 
 | | The Mouse PNGase-HR23 Complex Reveals a Complete Remodulation of the Protein-Protein Interface Compared to its Yeast Orthologs | | 分子名称: | CHLORIDE ION, PHQ-VAL-ALA-ASP-CF0, XP-C repair complementing complex 58 kDa protein, ... | | 著者 | Zhao, G, Zhou, X, Wang, L, Kisker, C, Lennarz, W.J, Schindelin, H. | | 登録日 | 2005-11-23 | | 公開日 | 2006-03-07 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Structure of the mouse peptide N-glycanase-HR23 complex suggests co-evolution of the endoplasmic reticulum-associated degradation and DNA repair pathways.
J.Biol.Chem., 281, 2006
|
|
2NRZ
 
 | |
2NRT
 
 | |
2NRW
 
 | |
2NRX
 
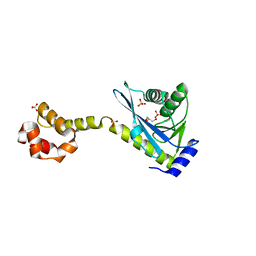 | | Crystal structure of the C-terminal half of UvrC, in the presence of sulfate molecules | | 分子名称: | GLYCEROL, SULFATE ION, UvrABC system protein C | | 著者 | Karakas, E, Truglio, J.J, Kisker, C. | | 登録日 | 2006-11-02 | | 公開日 | 2007-02-06 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of the C-terminal half of UvrC reveals an RNase H endonuclease domain with an Argonaute-like catalytic triad.
Embo J., 26, 2007
|
|
2NRV
 
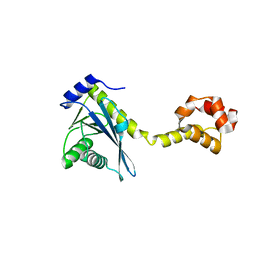 | |
2NRR
 
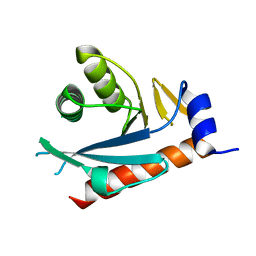 | |
4ALK
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 5-ethyl- 2-phenoxyphenol | | 分子名称: | 5-ETHYL-2-PHENOXYPHENOL, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, ... | | 著者 | Schiebel, J, Chang, A, Tonge, P.J, Kisker, C. | | 登録日 | 2012-03-04 | | 公開日 | 2012-05-09 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Staphylococcus Aureus Fabi: Inhibition, Substrate Recognition and Potential Implications for in Vivo Essentiality
Structure, 20, 2012
|
|
4A15
 
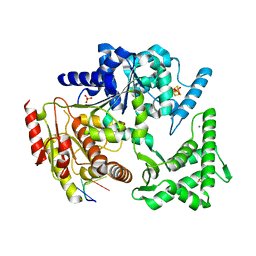 | | Crystal structure of an XPD DNA complex | | 分子名称: | 5'-D(*DTP*AP*CP*GP)-3', ATP-DEPENDENT DNA HELICASE TA0057, CALCIUM ION, ... | | 著者 | Kuper, J, Wolski, S.C, Michels, G, Kisker, C. | | 登録日 | 2011-09-14 | | 公開日 | 2012-02-01 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Functional and Structural Studies of the Nucleotide Excision Repair Helicase Xpd Suggest a Polarity for DNA Translocation.
Embo J., 31, 2011
|
|
