5E5D
 
 | |
5CY7
 
 | | Structure of Xoo1075, a peptide deformylase from Xanthomonas oryze pv oryze, in complex with fragment 275 | | 分子名称: | 2-(difluoromethyl)-1H-benzimidazole, ACETATE ION, CADMIUM ION, ... | | 著者 | Ngo, H.P.T, Kang, L.W. | | 登録日 | 2015-07-30 | | 公開日 | 2016-08-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure of Xoo1075, a peptide deformylase from Xanthomonas oryze pv oryze, in complex with fragment 275
To Be Published
|
|
5CX0
 
 | | Structure of Xoo1075, a peptide deformylase from Xanthomonas oryzae pv. oryzae, in complex with fragment 322 | | 分子名称: | 5-(propan-2-yl)-2-thioxodihydropyrimidine-4,6(1H,5H)-dione, ACETATE ION, CADMIUM ION, ... | | 著者 | Ngo, H.P.T, Kang, L.W. | | 登録日 | 2015-07-28 | | 公開日 | 2016-08-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure of Xoo1075, a peptide deformylase from Xanthomonas oryzae pv. oryzae, in complex with fragment 322
To Be Published
|
|
5CXJ
 
 | | Structure of Xoo1075, a peptide deformylase from Xanthomonas oryzae pv oryzae, in complex with fragment 124 | | 分子名称: | 2-(furan-2-yl)ethanamine, ACETATE ION, CADMIUM ION, ... | | 著者 | Ngo, H.P.T, Kang, L.W. | | 登録日 | 2015-07-29 | | 公開日 | 2016-08-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Structure of Xoo1075, a peptide deformylase from Xanthomonas oryzae pv oryzae, in complex with fragment 124
To Be Published
|
|
5CY8
 
 | | Structure of Xoo1075, a peptide deformylase from Xanthomonas oryzae pv oryze, in complex with fragment 244 | | 分子名称: | (3R)-2,3-dihydro[1,3]thiazolo[3,2-a]benzimidazol-3-ol, ACETATE ION, CADMIUM ION, ... | | 著者 | Ngo, H.P.T, Kang, L.W. | | 登録日 | 2015-07-30 | | 公開日 | 2016-08-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Structure of Xoo1075, a peptide deformylase from Xanthomonas oryzae pv oryze, in complex with fragment 244
To Be Published
|
|
5DMX
 
 | |
5C1O
 
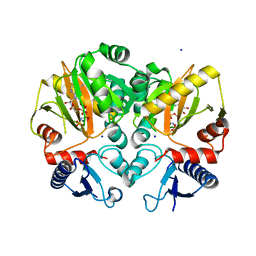 | | Crystal structure of AMP-PNP complexed D-alanine-D-alanine ligase(DDL) from Yersinia pestis | | 分子名称: | D-alanine--D-alanine ligase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | Tran, H.T, Kang, L.W, Hong, M.K. | | 登録日 | 2015-06-15 | | 公開日 | 2016-03-02 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure of D-alanine-D-alanine ligase from Yersinia pestis: nucleotide phosphate recognition by the serine loop.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4FXB
 
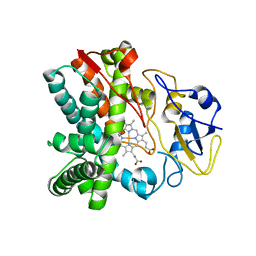 | | Crystal structure of CYP105N1 from Streptomyces coelicolor: a cytochrome P450 oxidase in the coelibactin siderophore biosynthetic pathway | | 分子名称: | PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 | | 著者 | Hong, M.K, Lim, Y.R, Kim, J.K, Kim, D.H, Kang, L.W. | | 登録日 | 2012-07-03 | | 公開日 | 2012-11-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of cytochrome P450 CYP105N1 from Streptomyces coelicolor, an oxidase in the coelibactin siderophore biosynthetic pathway
Arch.Biochem.Biophys., 528, 2012
|
|
7BZ4
 
 | |
7BYQ
 
 | |
7BZ1
 
 | |
7BZI
 
 | |
7BZ3
 
 | |
6ILA
 
 | | Two Glycerol complexed Crystal structure of fructuronate-tagaturonate epimerase UxaE from Cohnella laeviribosi | | 分子名称: | Fructuronate-tagaturonate epimerase UxaE, GLYCEROL, PHOSPHATE ION, ... | | 著者 | Choi, M.Y, Kang, L.W, Ho, T.H, Nguyen, D.Q, Lee, I.H, Lee, J.H, Park, Y.S, Park, H.J. | | 登録日 | 2018-10-17 | | 公開日 | 2019-10-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Crystal structure of fructuronate-tagaturonate epimerase UxaE from Cohnella laeviribosi
To be published
|
|
6IKT
 
 | |
6IL0
 
 | |
6IL9
 
 | | One Glycerol complexed Crystal structure of fructuronate-tagaturonate epimerase UxaE from Cohnella laeviribosi | | 分子名称: | Fructuronate-tagaturonate epimerase UxaE from Cohnella laeviribosi in complex with 1 glycerol, GLYCEROL, ZINC ION | | 著者 | Choi, M.Y, Kang, L.W, Ho, T.H, Nguyen, D.Q, Lee, I.H, Lee, J.H, Park, Y.S, Park, H.J. | | 登録日 | 2018-10-17 | | 公開日 | 2019-10-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.72005355 Å) | | 主引用文献 | Crystal structure of fructuronate-tagaturonate epimerase UxaE from Cohnella laeviribosi
To Be Published
|
|
6JFR
 
 | |
6J4N
 
 | | Structure of papua new guinea MBL-1(PNGM-1) native | | 分子名称: | Metallo-beta-lactamases PNGM-1, ZINC ION | | 著者 | Hong, M.K, Park, K.S, Jeon, J.H, Lee, J.H, Park, Y.S, Lee, S.H, Kang, L.W. | | 登録日 | 2019-01-10 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The novel metallo-beta-lactamase PNGM-1 from a deep-sea sediment metagenome: crystallization and X-ray crystallographic analysis.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6JF8
 
 | |
6JFS
 
 | |
6JET
 
 | |
6JF6
 
 | |
6JFO
 
 | |
6JF9
 
 | |
