5HMH
 
 | | HDM2 in complex with a 3,3-Disubstituted Piperidine | | 分子名称: | 4-[2-(4-{[(2R,3S)-2-propyl-1-{[4-(trifluoromethyl)pyridin-3-yl]carbonyl}-3-{[5-(trifluoromethyl)thiophen-3-yl]oxy}piperidin-3-yl]carbonyl}piperazin-1-yl)phenoxy]butanoic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | 著者 | Scapin, G. | | 登録日 | 2016-01-16 | | 公開日 | 2016-04-06 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Discovery of Novel 3,3-Disubstituted Piperidines as Orally Bioavailable, Potent, and Efficacious HDM2-p53 Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
7T5H
 
 | | Structure of rabies virus phosphoprotein C-terminal domain, wild type | | 分子名称: | 1,2-ETHANEDIOL, PHOSPHATE ION, Phosphoprotein, ... | | 著者 | Zhan, J, Metcalfe, R.D, Gooley, P.R, Griffin, M.D.W. | | 登録日 | 2021-12-12 | | 公開日 | 2022-04-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Molecular Basis of Functional Effects of Phosphorylation of the C-Terminal Domain of the Rabies Virus P Protein.
J.Virol., 96, 2022
|
|
7T5G
 
 | | Structure of rabies virus phosphoprotein C-terminal domain, S210E mutant | | 分子名称: | Phosphoprotein, SULFATE ION | | 著者 | Zhan, J, Metcalfe, R.D, Gooley, P.R, Griffin, M.D.W. | | 登録日 | 2021-12-12 | | 公開日 | 2022-04-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Molecular Basis of Functional Effects of Phosphorylation of the C-Terminal Domain of the Rabies Virus P Protein.
J.Virol., 96, 2022
|
|
7UMX
 
 | | Crystal structure of Acinetobacter baumannii FabI in complex with NAD and (R,E)-3-(7-amino-8-oxo-6,7,8,9-tetrahydro-5H-pyrido[2,3-b]azepin-3-yl)-N-methyl-N-((3-methylbenzofuran-2-yl)methyl)acrylamide | | 分子名称: | (2E)-3-[(7R)-7-amino-8-oxo-6,7,8,9-tetrahydro-5H-pyrido[2,3-b]azepin-3-yl]-N-methyl-N-[(3-methyl-1-benzofuran-2-yl)methyl]prop-2-enamide, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Hajian, B. | | 登録日 | 2022-04-08 | | 公開日 | 2023-02-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | An Iterative Approach Guides Discovery of the FabI Inhibitor Fabimycin, a Late-Stage Antibiotic Candidate with In Vivo Efficacy against Drug-Resistant Gram-Negative Infections
Acs Cent.Sci., 8, 2022
|
|
7UM8
 
 | | Crystal structure of E. Coli FabI in complex with NAD and (R,E)-3-(7-amino-8-oxo-6,7,8,9-tetrahydro-5H-pyrido[2,3-b]azepin-3-yl)-N-methyl-N-((3-methylbenzofuran-2-yl)methyl)acrylamide | | 分子名称: | (2E)-3-[(7R)-7-amino-8-oxo-6,7,8,9-tetrahydro-5H-pyrido[2,3-b]azepin-3-yl]-N-methyl-N-[(3-methyl-1-benzofuran-2-yl)methyl]prop-2-enamide, Enoyl-[acyl-carrier-protein] reductase [NADH] FabI, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Hajian, B. | | 登録日 | 2022-04-06 | | 公開日 | 2023-02-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | An Iterative Approach Guides Discovery of the FabI Inhibitor Fabimycin, a Late-Stage Antibiotic Candidate with In Vivo Efficacy against Drug-Resistant Gram-Negative Infections
Acs Cent.Sci., 8, 2022
|
|
7UMY
 
 | | Crystal structure of Acinetobacter baumannii FabI in complex with NAD and Fabimycin ((S,E)-3-(7-amino-8-oxo-6,7,8,9-tetrahydro-5H-pyrido[2,3-b]azepin-3-yl)-N-methyl-N-((3-methylbenzofuran-2-yl)methyl)acrylamide) | | 分子名称: | (2E)-3-[(7S)-7-amino-8-oxo-6,7,8,9-tetrahydro-5H-pyrido[2,3-b]azepin-3-yl]-N-methyl-N-[(3-methyl-1-benzofuran-2-yl)methyl]prop-2-enamide, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Hajian, B. | | 登録日 | 2022-04-08 | | 公開日 | 2023-02-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.74 Å) | | 主引用文献 | An Iterative Approach Guides Discovery of the FabI Inhibitor Fabimycin, a Late-Stage Antibiotic Candidate with In Vivo Efficacy against Drug-Resistant Gram-Negative Infections
Acs Cent.Sci., 8, 2022
|
|
4PFK
 
 | |
1ZKG
 
 | |
1ZX8
 
 | |
1ZCZ
 
 | |
6KUW
 
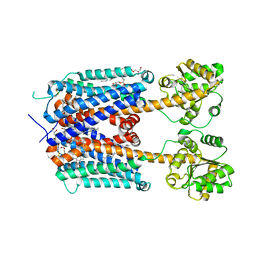 | | Crystal structure of human alpha2C adrenergic G protein-coupled receptor. | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (8~{a}~{R},12~{a}~{S},13~{a}~{R})-12-ethylsulfonyl-3-methoxy-5,6,8,8~{a},9,10,11,12~{a},13,13~{a}-decahydroisoquinolino[2,1-g][1,6]naphthyridine, Alpha-2C adrenergic receptor, ... | | 著者 | Chen, X.Y, Wu, L.J, Wu, D, Zhong, G.S. | | 登録日 | 2019-09-02 | | 公開日 | 2019-12-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of human alpha2C adrenergic G protein-coupled receptor.
To Be Published
|
|
2A6A
 
 | |
4XJ8
 
 | |
4XHB
 
 | |
4XJW
 
 | |
4XJ9
 
 | |
4XJZ
 
 | |
4XOG
 
 | |
4XIO
 
 | |
4XJU
 
 | | Crystal structure of the NanB sialidase from streptococcus pneumoniae in complex with 4-acetamido-2-fluoro-3-hydroxy-6-[1,2-dihydroxyethyl]-7,8-dioxabicyclo[3.2.1]octane-1-carboxylic acid | | 分子名称: | (1R,2R,3R,4R,5R,7R)-2-(acetylamino)-7-[(1R)-1,2-dihydroxyethyl]-4-fluoro-3-hydroxy-6,8-dioxabicyclo[3.2.1]octane-5-carboxylic acid, DIMETHYL SULFOXIDE, PHOSPHATE ION, ... | | 著者 | Brear, P. | | 登録日 | 2015-01-09 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | `The Hunt for Serendipitous Allosteric Sites: Discovery of a novel allosteric inhibitor of the bacterial sialidase NanB
To be published
|
|
4XHX
 
 | |
4XIK
 
 | |
4XJA
 
 | | Crystal structure of the NanB sialidase from streptococcus pneumoniae in complex with 5-acetamido-2,3-difluoro-3-hydroxy-6-[1,2,3-trihydroxypropyl]oxane-2-carboxylic acid | | 分子名称: | (1s,3R,4S)-1-[(cyclohexylamino)methyl]-3,4-dihydroxycyclopentanesulfonic acid, (2R,3R,4R,5R,6R)-5-acetamido-2,3-difluoro-4-hydroxy-6-[(1R,2R)-1,2,3-trihydroxypropyl]tetrahydro-2H-pyran-2-carboxylic acid, DIMETHYL SULFOXIDE, ... | | 著者 | Brear, P. | | 登録日 | 2015-01-08 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | `The Hunt for Serendipitous Allosteric Sites: Discovery of a novel allosteric inhibitor of the bacterial sialidase NanB
To be published
|
|
4XIL
 
 | |
4XMA
 
 | |
