6O2Y
 
 | | Crystal structure of IDH1 R132H mutant in complex with compound 24 | | 分子名称: | 4-{[(6-chloro-2-oxo-1,2-dihydroquinolin-3-yl)methyl]amino}-2-methoxybenzonitrile, BETA-MERCAPTOETHANOL, Isocitrate dehydrogenase [NADP] cytoplasmic, ... | | 著者 | Toms, A.V, Lin, J. | | 登録日 | 2019-02-25 | | 公開日 | 2019-06-26 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Discovery and Optimization of Quinolinone Derivatives as Potent, Selective, and Orally Bioavailable Mutant Isocitrate Dehydrogenase 1 (mIDH1) Inhibitors.
J.Med.Chem., 62, 2019
|
|
4DK5
 
 | | Crystal structure of human PI3K-gamma in complex with a pyridyl-triazine inhibitor | | 分子名称: | 4-(2-[(6-methoxypyridin-3-yl)amino]-5-{[4-(methylsulfonyl)piperazin-1-yl]methyl}pyridin-3-yl)-6-methyl-1,3,5-triazin-2-amine, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | 著者 | Whittington, D.A, Tang, J, Yakowec, P. | | 登録日 | 2012-02-03 | | 公開日 | 2012-05-16 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Structure-based design of a novel series of potent, selective inhibitors of the class I phosphatidylinositol 3-kinases.
J.Med.Chem., 55, 2012
|
|
4CFX
 
 | | Structure-based design of C8-substituted O6-cyclohexylmethoxyguanine CDK1 and 2 inhibitors. | | 分子名称: | 3-[2-amino-6-(cyclohexylmethoxy)-7H-purin-8-yl]benzenesulfonamide, CYCLIN-A2, CYCLIN-DEPENDENT KINASE 2 | | 著者 | Carbain, B, Paterson, D.J, Anscombe, E, Campbell, A, Cano, C, Echalier, A, Endicott, J, Golding, B.T, Haggerty, K, Hardcastle, I.R, Jewsbury, P, Newell, D.R, Noble, M.E.M, Roche, C, Wang, L.Z, Griffin, R. | | 登録日 | 2013-11-19 | | 公開日 | 2014-12-10 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | 8-Substituted O6-Cyclohexylmethylguanine Cdk2 Inhibitors; Using Structure-Based Inhibitor Design to Optimise an Alternative Binding Mode.
J.Med.Chem., 57, 2014
|
|
8WXL
 
 | | Structure of the SARS-CoV-2 BA.2.86 spike glycoprotein (closed state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | 登録日 | 2023-10-30 | | 公開日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (2.59 Å) | | 主引用文献 | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1
Nat Commun, 2024
|
|
8XUX
 
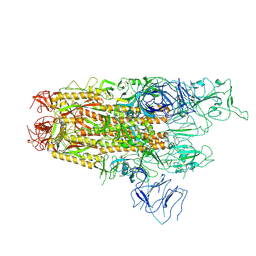 | | Structure of the SARS-CoV-2 BA.2.86 spike protein (1-up state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | 登録日 | 2024-01-14 | | 公開日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.22 Å) | | 主引用文献 | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1
Nat Commun, 2024
|
|
8XV0
 
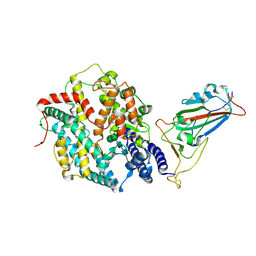 | | Structure of SARS-CoV-2 BA.2.86 spike RBD in complex with ACE2 (up state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | 登録日 | 2024-01-14 | | 公開日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1
Nat Commun, 2024
|
|
8XV1
 
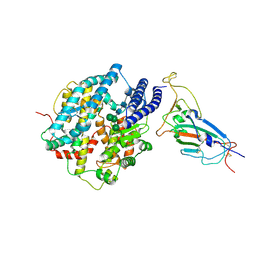 | | Structure of SARS-CoV-2 BA.2.86 spike RBD in complex with ACE2 (down state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | 登録日 | 2024-01-14 | | 公開日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.05 Å) | | 主引用文献 | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1
Nat Commun, 2024
|
|
9IU1
 
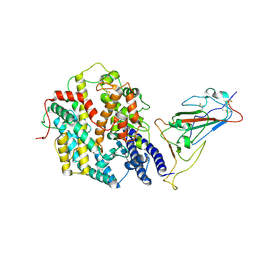 | | Structure of SARS-CoV-2 JN.1 spike RBD in complex with ACE2 (up state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | 登録日 | 2024-07-20 | | 公開日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1
Nat Commun, 2024
|
|
8U56
 
 | |
8U5C
 
 | |
8U5N
 
 | |
8U54
 
 | |
8U5V
 
 | |
8U58
 
 | |
8U5X
 
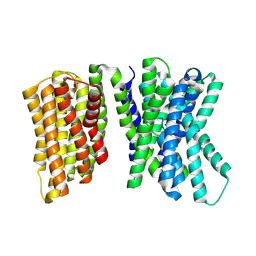 | |
8U5Q
 
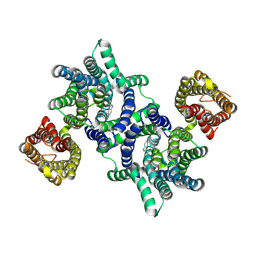 | |
5GGZ
 
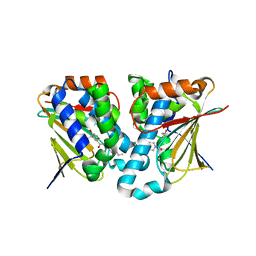 | | Crystal structure of novel inhibitor bound with Hsp90 | | 分子名称: | Heat shock protein HSP 90-alpha, [2,4-bis(oxidanyl)-5-propan-2-yl-phenyl]-(2-ethoxy-7,8-dihydro-5~{H}-pyrido[4,3-d]pyrimidin-6-yl)methanone | | 著者 | Chen, T.T, Li, J, Xu, Y.C. | | 登録日 | 2016-06-16 | | 公開日 | 2017-03-08 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.015 Å) | | 主引用文献 | Novel Tetrahydropyrido[4,3-d]pyrimidines as Potent Inhibitors of Chaperone Heat Shock Protein 90
J. Med. Chem., 59, 2016
|
|
8XUY
 
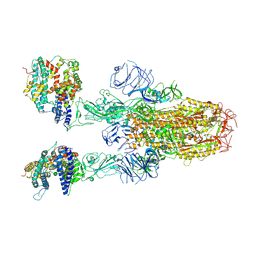 | | Structure of SARS-CoV-2 BA.2.86 spike glycoprotein in complex with ACE2 (2-up state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | 登録日 | 2024-01-14 | | 公開日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.14 Å) | | 主引用文献 | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1
Nat Commun, 2024
|
|
8XUZ
 
 | | Structure of SARS-CoV-2 BA.2.86 spike glycoprotein in complex with ACE2 (2-up and 1-down state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | 登録日 | 2024-01-14 | | 公開日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.05 Å) | | 主引用文献 | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1
Nat Commun, 2024
|
|
8XVM
 
 | | Structure of SARS-CoV-2 BA.2.86 spike glycoprotein in complex with ACE2 (3-up state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | 登録日 | 2024-01-15 | | 公開日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (2.77 Å) | | 主引用文献 | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1
Nat Commun, 2024
|
|
7WG7
 
 | | Acidic Omicron Spike Trimer | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Cui, Z. | | 登録日 | 2021-12-28 | | 公開日 | 2022-06-22 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron.
Cell, 185, 2022
|
|
7WGC
 
 | | Neutral Omicron Spike Trimer in complex with ACE2. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | 著者 | Cui, Z. | | 登録日 | 2021-12-28 | | 公開日 | 2022-06-22 | | 最終更新日 | 2022-07-13 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron.
Cell, 185, 2022
|
|
7WG9
 
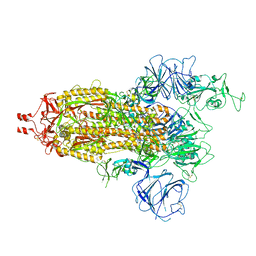 | | Delta Spike Trimer(1 RBD Up) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Cui, Z. | | 登録日 | 2021-12-28 | | 公開日 | 2022-06-22 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron.
Cell, 185, 2022
|
|
7WGB
 
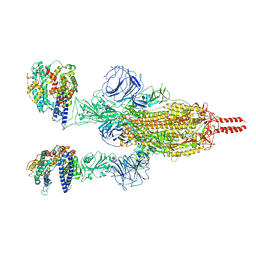 | | Neutral Omicron Spike Trimer in complex with ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Cui, Z. | | 登録日 | 2021-12-28 | | 公開日 | 2022-06-22 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron.
Cell, 185, 2022
|
|
7WG8
 
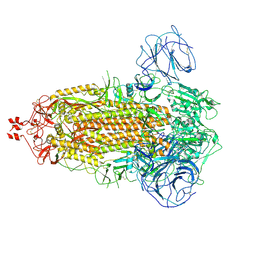 | | Delta Spike Trimer(3 RBD Down) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Cui, Z. | | 登録日 | 2021-12-28 | | 公開日 | 2022-06-22 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron.
Cell, 185, 2022
|
|
