4GFT
 
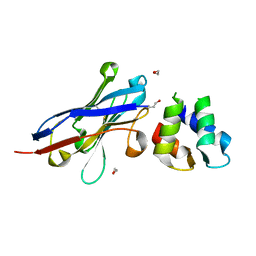 | | Malaria invasion machinery protein-Nanobody complex | | 分子名称: | 1,2-ETHANEDIOL, Myosin A tail domain interacting protein, Nanobody | | 著者 | Khamrui, S, Turley, S, Pardon, E, Steyaert, J, Verlinde, C, Fan, E, Bergman, L.W, Hol, W.G.J. | | 登録日 | 2012-08-03 | | 公開日 | 2013-07-03 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The structure of the D3 domain of Plasmodium falciparum myosin tail interacting protein MTIP in complex with a nanobody.
Mol.Biochem.Parasitol., 190, 2013
|
|
4DAY
 
 | |
5NX0
 
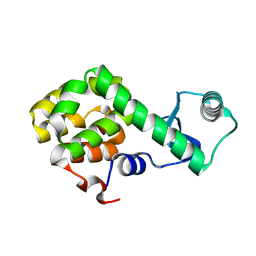 | | Structure of Spin-labelled T4 lysozyme mutant L115C-R119C-R1 at room temperature | | 分子名称: | Endolysin | | 著者 | Gohlke, U, Loll, B, Consentius, P, Mueller, R, Kaupp, M, Heinemann, U, Wahl, M.C, Risse, T. | | 登録日 | 2017-05-09 | | 公開日 | 2017-07-19 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.803 Å) | | 主引用文献 | Combining EPR spectroscopy and X-ray crystallography to elucidate the structure and dynamics of conformationally constrained spin labels in T4 lysozyme single crystals.
Phys Chem Chem Phys, 19, 2017
|
|
2Z9A
 
 | |
2HSG
 
 | |
2LDL
 
 | | Solution NMR Structure of the HIV-1 Exon Splicing Silencer 3 | | 分子名称: | RNA (27-MER) | | 著者 | Mishler, C, Levengood, J.D, Johnson, C.A, Rajan, P, Znosko, B.M. | | 登録日 | 2011-05-27 | | 公開日 | 2011-12-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of the HIV-1 Exon Splicing Silencer 3.
J.Mol.Biol., 415, 2012
|
|
2MQH
 
 | |
1AUW
 
 | |
1AVO
 
 | |
2LBM
 
 | |
1DZT
 
 | | RMLC FROM SALMONELLA TYPHIMURIUM | | 分子名称: | 3'-O-ACETYLTHYMIDINE-(5' DIPHOSPHATE PHENYL ESTER), 3'-O-ACETYLTHYMIDINE-5'-DIPHOSPHATE, DTDP-4-DEHYDRORHAMNOSE 3,5-EPIMERASE, ... | | 著者 | Naismith, J.H, Giraud, M.F. | | 登録日 | 2000-03-07 | | 公開日 | 2000-04-18 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Rmlc, the Third Enzyme of Dtdp-L-Rhamnose Pathway, is a New Class of Epimerase.
Nat.Struct.Biol., 7, 2000
|
|
1DPI
 
 | |
1DZR
 
 | | RmlC from Salmonella typhimurium | | 分子名称: | DTDP-4-DEHYDRORHAMNOSE 3,5-EPIMERASE, GLYCEROL, SULFATE ION | | 著者 | Naismith, J.H, Giraud, M.F. | | 登録日 | 2000-03-07 | | 公開日 | 2000-04-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Rmlc, the Third Enzyme of Dtdp-L-Rhamnose Pathway, is a New Class of Epimerase.
Nat.Struct.Biol., 7, 2000
|
|
1D62
 
 | |
1D61
 
 | | THE STRUCTURE OF THE B-DNA DECAMER C-C-A-A-C-I-T-T-G-G: MONOCLINIC FORM | | 分子名称: | CACODYLATE ION, CALCIUM ION, DNA (5'-D(*CP*CP*AP*AP*CP*IP*TP*TP*GP*G)-3') | | 著者 | Lipanov, A, Kopka, M.L, Kaczor-Grzeskowiak, M, Quintana, J, Dickerson, R.E. | | 登録日 | 1992-02-26 | | 公開日 | 1993-04-15 | | 最終更新日 | 2023-07-26 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structure of the B-DNA decamer C-C-A-A-C-I-T-T-G-G in two different space groups: conformational flexibility of B-DNA.
Biochemistry, 32, 1993
|
|
1AMF
 
 | | CRYSTAL STRUCTURE OF MODA, A MOLYBDATE TRANSPORT PROTEIN, COMPLEXED WITH MOLYBDATE | | 分子名称: | MOLYBDATE ION, MOLYBDATE TRANSPORT PROTEIN MODA | | 著者 | Hu, Y, Rech, S, Gunsalus, R.P, Rees, D.C. | | 登録日 | 1997-06-13 | | 公開日 | 1997-12-24 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal structure of the molybdate binding protein ModA.
Nat.Struct.Biol., 4, 1997
|
|
