4XP8
 
 | |
7TC5
 
 | | All Phe-Azurin variant - F15Y | | 分子名称: | Azurin, COPPER (II) ION, NITRATE ION, ... | | 著者 | Fedoretz-Maxwell, B.P, Worrall, L.J, Strynadka, N.C.J, Warren, J.J. | | 登録日 | 2021-12-22 | | 公開日 | 2022-06-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | The Impact of Second Coordination Sphere Methionine-Aromatic Interactions in Copper Proteins.
Inorg.Chem., 61, 2022
|
|
7TC6
 
 | | All Phe-Azurin variant - F15W | | 分子名称: | Azurin, COPPER (II) ION, NITRATE ION | | 著者 | Fedoretz-Maxwell, B.P, Worrall, L.J, Strynadka, N.C.J, Warren, J.J. | | 登録日 | 2021-12-22 | | 公開日 | 2022-06-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The Impact of Second Coordination Sphere Methionine-Aromatic Interactions in Copper Proteins.
Inorg.Chem., 61, 2022
|
|
7UZ2
 
 | | Structure of beta-glycosidase from Sulfolobus solfataricus in complex with C5a-fluoro-valienide. | | 分子名称: | (1R,2S,3R,4R)-5-fluoro-6-(hydroxymethyl)cyclohex-5-ene-1,2,3,4-tetrol, Beta-galactosidase | | 著者 | Danby, P.M, Jeong, A, Sim, L, Sweeney, R.P, Wardman, J.F, Geissner, A, Worrall, L.J, Strynadka, N.C.J, Withers, S.G. | | 登録日 | 2022-05-08 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Vinyl Halide-Modified Unsaturated Cyclitols are Mechanism-Based Glycosidase Inhibitors.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7UZ1
 
 | | Structure of beta-glycosidase from Sulfolobus solfataricus in complex with C5a-bromo-valienide. | | 分子名称: | (1R,2S,3R,4R)-5-bromo-6-(hydroxymethyl)cyclohex-5-ene-1,2,3,4-tetrol, 1,2-ETHANEDIOL, Beta-galactosidase | | 著者 | Danby, P.M, Jeong, A, Sim, L, Sweeney, R.P, Wardman, J.F, Karimi, R, Geissner, A, Worrall, L.J, Strynadka, N.C.J, Withers, S.G. | | 登録日 | 2022-05-08 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Vinyl Halide-Modified Unsaturated Cyclitols are Mechanism-Based Glycosidase Inhibitors.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6PEP
 
 | | Focussed refinement of InvGN0N1:SpaPQR:PrgIJ from the Salmonella SPI-1 injectisome needle complex | | 分子名称: | Protein InvG, Protein PrgH, Protein PrgI, ... | | 著者 | Hu, J, Worrall, L.J, Strynadka, N.C.J. | | 登録日 | 2019-06-20 | | 公開日 | 2019-10-23 | | 最終更新日 | 2020-01-08 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | T3S injectisome needle complex structures in four distinct states reveal the basis of membrane coupling and assembly.
Nat Microbiol, 4, 2019
|
|
6PEE
 
 | |
6PEM
 
 | | Focussed refinement of InvGN0N1:SpaPQR:PrgHK from Salmonella SPI-1 injectisome NC-base | | 分子名称: | Lipoprotein PrgK, Protein InvG, Protein PrgH, ... | | 著者 | Hu, J, Worrall, L.J, Strynadka, N.C.J. | | 登録日 | 2019-06-20 | | 公開日 | 2019-10-23 | | 最終更新日 | 2020-01-08 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | T3S injectisome needle complex structures in four distinct states reveal the basis of membrane coupling and assembly.
Nat Microbiol, 4, 2019
|
|
6Q15
 
 | | Structure of the Salmonella SPI-1 injectisome needle complex | | 分子名称: | Lipoprotein PrgK, Protein InvG, Protein PrgH, ... | | 著者 | Hu, J, Worrall, L.J, Strynadka, N.C.J. | | 登録日 | 2019-08-02 | | 公開日 | 2019-10-23 | | 最終更新日 | 2020-01-08 | | 実験手法 | ELECTRON MICROSCOPY (5.15 Å) | | 主引用文献 | T3S injectisome needle complex structures in four distinct states reveal the basis of membrane coupling and assembly.
Nat Microbiol, 4, 2019
|
|
6Q14
 
 | | Structure of the Salmonella SPI-1 injectisome NC-base | | 分子名称: | Lipoprotein PrgK, Protein InvG, Protein PrgH, ... | | 著者 | Hu, J, Worrall, L.J, Strynadka, N.C.J. | | 登録日 | 2019-08-02 | | 公開日 | 2019-10-23 | | 最終更新日 | 2020-01-08 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | T3S injectisome needle complex structures in four distinct states reveal the basis of membrane coupling and assembly.
Nat Microbiol, 4, 2019
|
|
6Q16
 
 | | Focussed refinement of InvGN0N1:PrgHK:SpaPQR:PrgIJ from Salmonella SPI-1 injectisome NC-base | | 分子名称: | Lipoprotein PrgK, Protein InvG, Protein PrgH, ... | | 著者 | Hu, J, Worrall, L.J, Strynadka, N.C.J. | | 登録日 | 2019-08-02 | | 公開日 | 2019-10-23 | | 最終更新日 | 2020-01-15 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | T3S injectisome needle complex structures in four distinct states reveal the basis of membrane coupling and assembly.
Nat Microbiol, 4, 2019
|
|
8TCR
 
 | | Structure of glucose bound Alistipes sp. 3-Keto-beta-glucopyranoside-1,2-Lyase AL1 | | 分子名称: | COBALT (II) ION, MALONATE ION, Sugar phosphate isomerase, ... | | 著者 | Lazarski, A.C, Worrall, L.J, Strynadka, N.C.J. | | 登録日 | 2023-07-02 | | 公開日 | 2024-06-12 | | 最終更新日 | 2024-07-17 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | An alternative broad-specificity pathway for glycan breakdown in bacteria.
Nature, 631, 2024
|
|
8TCT
 
 | | Structure of 3K-GlcH bound Bacteroides thetaiotaomicron 3-Keto-beta-glucopyranoside-1,2-Lyase BT1 | | 分子名称: | 1,5-anhydro-D-ribo-hex-3-ulose, COBALT (II) ION, PHOSPHATE ION, ... | | 著者 | Lazarski, A.C, Worrall, L.J, Strynadka, N.C.J. | | 登録日 | 2023-07-02 | | 公開日 | 2024-06-12 | | 最終更新日 | 2024-07-17 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | An alternative broad-specificity pathway for glycan breakdown in bacteria.
Nature, 631, 2024
|
|
8TDE
 
 | |
8TCD
 
 | | Structure of Alistipes sp. 3-Keto-beta-glucopyranoside-1,2-Lyase AL1 | | 分子名称: | ACETATE ION, COBALT (II) ION, GLYCEROL, ... | | 著者 | Lazarski, A.C, Worrall, L.J, Strynadka, N.C.J. | | 登録日 | 2023-06-30 | | 公開日 | 2024-06-12 | | 最終更新日 | 2024-07-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | An alternative broad-specificity pathway for glycan breakdown in bacteria.
Nature, 631, 2024
|
|
8TDA
 
 | |
8TDF
 
 | |
8TDI
 
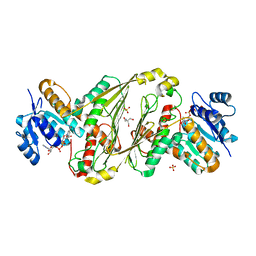 | | Structure of P2B11 Glucuronide-3-dehydrogenase | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, P2B11 Glucuronide-3-dehydrogenase, ... | | 著者 | Lazarski, A.C, Worrall, L.J, Strynadka, N.C.J. | | 登録日 | 2023-07-03 | | 公開日 | 2024-06-12 | | 最終更新日 | 2024-07-17 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | An alternative broad-specificity pathway for glycan breakdown in bacteria.
Nature, 631, 2024
|
|
8TCS
 
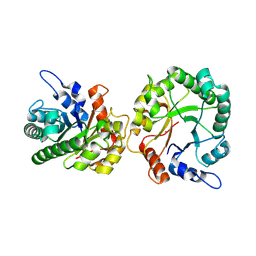 | | Structure of trehalose bound Alistipes sp. 3-Keto-beta-glucopyranoside-1,2-Lyase AL1 | | 分子名称: | ACETATE ION, COBALT (II) ION, Xylose isomerase-like TIM barrel domain-containing protein, ... | | 著者 | Lazarski, A.C, Worrall, L.J, Strynadka, N.C.J. | | 登録日 | 2023-07-02 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-07-17 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | An alternative broad-specificity pathway for glycan breakdown in bacteria.
Nature, 631, 2024
|
|
8TDH
 
 | |
3J1W
 
 | |
3J1X
 
 | | A refined model of the prototypical Salmonella typhimurium T3SS basal body reveals the molecular basis for its assembly | | 分子名称: | Protein PrgH | | 著者 | Sgourakis, N.G, Bergeron, J.R.C, Worrall, L.J, Strynadka, N.C.J, Baker, D. | | 登録日 | 2012-07-10 | | 公開日 | 2013-05-22 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (11.7 Å) | | 主引用文献 | A Refined Model of the Prototypical Salmonella SPI-1 T3SS Basal Body Reveals the Molecular Basis for Its Assembly.
Plos Pathog., 9, 2013
|
|
7KHP
 
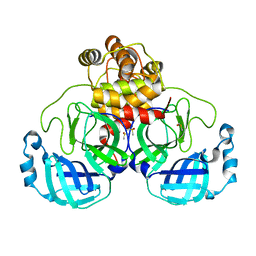 | | Acyl-enzyme intermediate structure of SARS-CoV-2 Mpro in complex with its C-terminal autoprocessing sequence. | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Lee, J, Worrall, L.J, Paetzel, M, Strynadka, N.C.J. | | 登録日 | 2020-10-21 | | 公開日 | 2020-10-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystallographic structure of wild-type SARS-CoV-2 main protease acyl-enzyme intermediate with physiological C-terminal autoprocessing site.
Nat Commun, 11, 2020
|
|
7JP1
 
 | |
7JOY
 
 | |
