1IGD
 
 | |
1E0J
 
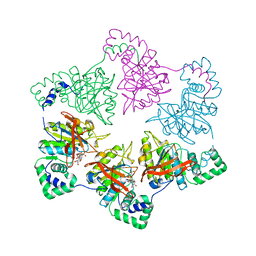 | | gp4d helicase from phage T7 ADPNP complex | | 分子名称: | DNA HELICASE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Singleton, M.R, Sawaya, M.R, Ellenberger, T, Wigley, D.B. | | 登録日 | 2000-03-30 | | 公開日 | 2000-06-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal Structure of T7 Gene 4 Ring Helicase Indicates a Mechanism for Sequential Hydrolysis of Nucleotides
Cell(Cambridge,Mass.), 101, 2000
|
|
1D0Q
 
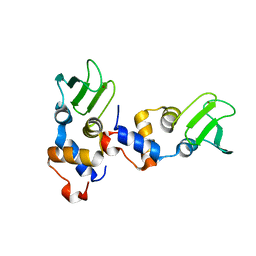 | |
1E0K
 
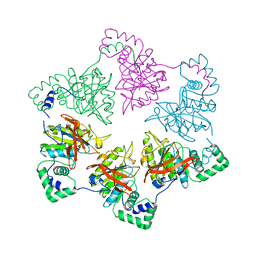 | | gp4d helicase from phage T7 | | 分子名称: | DNA HELICASE | | 著者 | Singleton, M.R, Sawaya, M.R, Ellenberger, T, Wigley, D.B. | | 登録日 | 2000-03-30 | | 公開日 | 2000-06-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Crystal Structure of T7 Gene 4 Ring Helicase Indicates a Mechanism for Sequential Hydrolysis of Nucleotides
Cell(Cambridge,Mass.), 101, 2000
|
|
1B04
 
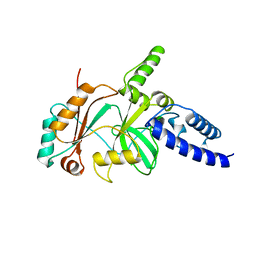 | |
1H2I
 
 | | Human Rad52 protein, N-terminal domain | | 分子名称: | DNA REPAIR PROTEIN RAD52 HOMOLOG | | 著者 | Singleton, M.R, Wentzell, L.M, Liu, Y, West, S.C, Wigley, D.B. | | 登録日 | 2002-08-09 | | 公開日 | 2002-10-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure of the Single-Strand Annealing Domain of Human Rad52 Protein
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1GM5
 
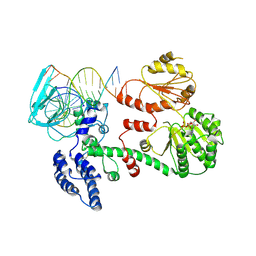 | | Structure of RecG bound to three-way DNA junction | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-(*CP*AP*GP*CP*TP*CP*CP*AP*TP*GP*AP*TP* CP*AP*TP*TP*GP*GP*CP*A)-3'), DNA (5'-(*GP*AP*GP*CP*AP*CP*TP*GP*C)-3'), ... | | 著者 | Singleton, M.R, Scaife, S, Wigley, D.B. | | 登録日 | 2001-09-11 | | 公開日 | 2001-10-03 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (3.24 Å) | | 主引用文献 | Structural Analysis of DNA Replication Fork Reversal by Recg
Cell(Cambridge,Mass.), 107, 2001
|
|
5OAF
 
 | | Human Rvb1/Rvb2 heterohexamer in INO80 complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, RuvB-like 1, RuvB-like 2 | | 著者 | Aramayo, R.J, Bythell-Douglas, R, Ayala, R, Willhoft, O, Wigley, D, Zhang, X. | | 登録日 | 2017-06-21 | | 公開日 | 2017-12-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4.06 Å) | | 主引用文献 | Cryo-EM structures of the human INO80 chromatin-remodeling complex.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3U44
 
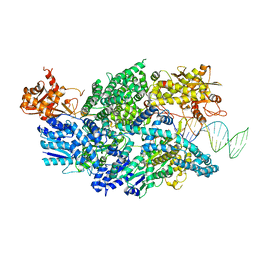 | | Crystal structure of AddAB-DNA complex | | 分子名称: | ATP-dependent helicase/deoxyribonuclease subunit B, ATP-dependent helicase/nuclease subunit A, DNA (36-MER), ... | | 著者 | Saikrishnan, K, Krajewski, W, Wigley, D. | | 登録日 | 2011-10-07 | | 公開日 | 2012-03-21 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.201 Å) | | 主引用文献 | Insights into Chi recognition from the structure of an AddAB-type helicase-nuclease complex.
Embo J., 31, 2012
|
|
3U4Q
 
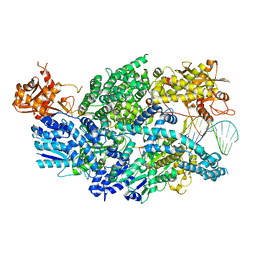 | | Structure of AddAB-DNA complex at 2.8 angstroms | | 分子名称: | 1,2-ETHANEDIOL, ATP-dependent helicase/deoxyribonuclease subunit B, ATP-dependent helicase/nuclease subunit A, ... | | 著者 | Saikrishnan, K, Krajewski, W, Wigley, D. | | 登録日 | 2011-10-10 | | 公開日 | 2012-03-21 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Insights into Chi recognition from the structure of an AddAB-type helicase-nuclease complex.
Embo J., 31, 2012
|
|
5DMA
 
 | |
6FWS
 
 | |
4CEH
 
 | | Crystal structure of AddAB with a forked DNA substrate | | 分子名称: | ATP-DEPENDENT HELICASE/DEOXYRIBONUCLEASE SUBUNIT B, ATP-DEPENDENT HELICASE/NUCLEASE SUBUNIT A, DNA, ... | | 著者 | Krajewski, W.W, Wilkinson, M, Fu, X, Cronin, N.B, Wigley, D. | | 登録日 | 2013-11-11 | | 公開日 | 2014-03-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.24 Å) | | 主引用文献 | Structural Basis for Translocation by Addab Helicase-Nuclease and its Arrest at Chi Sites.
Nature, 508, 2014
|
|
4CEI
 
 | | Crystal structure of ADPNP-bound AddAB with a forked DNA substrate | | 分子名称: | ATP-DEPENDENT HELICASE/DEOXYRIBONUCLEASE SUBUNIT B, ATP-DEPENDENT HELICASE/NUCLEASE SUBUNIT A, DNA, ... | | 著者 | Krajewski, W.W, Wilkinson, M, Fu, X, Cronin, N.B, Wigley, D. | | 登録日 | 2013-11-11 | | 公開日 | 2014-03-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural Basis for Translocation by Addab Helicase-Nuclease and its Arrest at Chi Sites.
Nature, 508, 2014
|
|
1AJ6
 
 | |
4CEJ
 
 | | Crystal structure of AddAB-DNA-ADPNP complex at 3 Angstrom resolution | | 分子名称: | ATP-DEPENDENT HELICASE/DEOXYRIBONUCLEASE SUBUNIT B, ATP-DEPENDENT HELICASE/NUCLEASE SUBUNIT A, DNA, ... | | 著者 | Krajewski, W.W, Wilkinson, M, Fu, X, Cronin, N.B, Wigley, D. | | 登録日 | 2013-11-11 | | 公開日 | 2014-03-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural Basis for Translocation by Addab Helicase-Nuclease and its Arrest at Chi Sites.
Nature, 508, 2014
|
|
1P7F
 
 | |
1P7E
 
 | |
1TA8
 
 | |
1TAE
 
 | |
