6IF7
 
 | | Crystal Structure of AA10 Lytic Polysaccharide Monooxygenase from Tectaria macrodonta | | 分子名称: | COPPER (II) ION, Chitin binding protein, GLYCEROL, ... | | 著者 | Archana, A, Yadav, S.K, Singh, P.K, Vasudev, P.G. | | 登録日 | 2018-09-18 | | 公開日 | 2019-04-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Insecticidal fern protein Tma12 is possibly a lytic polysaccharide monooxygenase.
Planta, 249, 2019
|
|
6KML
 
 | | 2.09 Angstrom resolution crystal structure of tetrameric HigBA toxin-antitoxin complex from E.coli | | 分子名称: | Antitoxin HigA, mRNA interferase toxin HigB | | 著者 | Jadhav, P, Sinha, V.K, Rothweiler, U, Singh, M. | | 登録日 | 2019-07-31 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.095 Å) | | 主引用文献 | 2.09 angstrom Resolution structure of E. coli HigBA toxin-antitoxin complex reveals an ordered DNA-binding domain and intrinsic dynamics in antitoxin.
Biochem.J., 477, 2020
|
|
6KMQ
 
 | | 2.3 Angstrom resolution structure of dimeric HigBA toxin-antitoxin complex from E. coli | | 分子名称: | Antitoxin HigA, mRNA interferase toxin HigB | | 著者 | Jadhav, P, Sinha, V.K, Rothweiler, U, Singh, M. | | 登録日 | 2019-07-31 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | 2.09 angstrom Resolution structure of E. coli HigBA toxin-antitoxin complex reveals an ordered DNA-binding domain and intrinsic dynamics in antitoxin.
Biochem.J., 477, 2020
|
|
7M05
 
 | |
5Z47
 
 | | Crystal structure of pyrrolidone carboxylate peptidase I with disordered loop A from Deinococcus radiodurans R1 | | 分子名称: | DIMETHYL SULFOXIDE, Pyrrolidone-carboxylate peptidase | | 著者 | Agrawal, R, Kumar, A, Kumar, A, Makde, R.D. | | 登録日 | 2018-01-10 | | 公開日 | 2019-01-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structures of pyrrolidone-carboxylate peptidase I from Deinococcus radiodurans reveal the mechanism of L-pyroglutamate recognition.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5ZAU
 
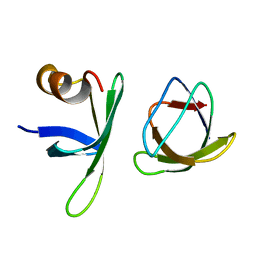 | |
5Z48
 
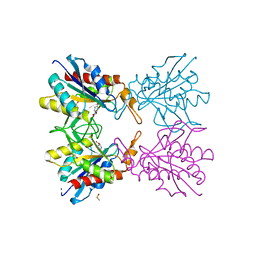 | | Crystal structure of pyrrolidone carboxylate peptidase I from Deinococcus radiodurans R1 bound to pyroglutamate | | 分子名称: | DIMETHYL SULFOXIDE, PYROGLUTAMIC ACID, Pyrrolidone-carboxylate peptidase, ... | | 著者 | Agrawal, R, Kumar, A, Kumar, A, Makde, R.D. | | 登録日 | 2018-01-10 | | 公開日 | 2019-01-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.551 Å) | | 主引用文献 | Crystal structures of pyrrolidone-carboxylate peptidase I from Deinococcus radiodurans reveal the mechanism of L-pyroglutamate recognition.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5L02
 
 | | S324T variant of B. pseudomallei KatG | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Catalase-peroxidase, PHOSPHATE ION, ... | | 著者 | Loewen, P.C. | | 登録日 | 2016-07-26 | | 公開日 | 2016-08-10 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural characterization of the Ser324Thr variant of the catalase-peroxidase (KatG) from Burkholderia pseudomallei
J. Mol. Biol., 345, 2005
|
|
6IFF
 
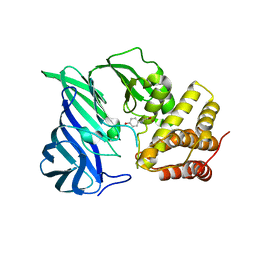 | | Crystal structure of M1 zinc metallopeptidase E323A mutant from Deinococcus radiodurans | | 分子名称: | SODIUM ION, TYROSINE, ZINC ION, ... | | 著者 | Agrawal, R, Kumar, A, Kumar, A, Gaur, N.K, Makde, R.D. | | 登録日 | 2018-09-20 | | 公開日 | 2019-09-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Structural basis for the unusual substrate specificity of unique two-domain M1 metallopeptidase.
Int.J.Biol.Macromol., 147, 2020
|
|
6KP1
 
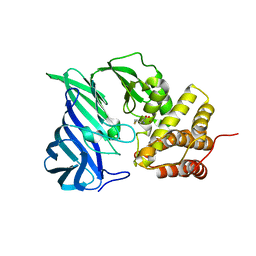 | | Crystal structure of two domain M1 zinc metallopeptidase E323A mutant bound to L-methionine amino acid | | 分子名称: | METHIONINE, SODIUM ION, ZINC ION, ... | | 著者 | Agrawal, R, Kumar, A, Kumar, A, Makde, R.D. | | 登録日 | 2019-08-13 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Structural basis for the unusual substrate specificity of unique two-domain M1 metallopeptidase.
Int.J.Biol.Macromol., 147, 2020
|
|
6KP0
 
 | | Crystal structure of two domain M1 zinc metallopeptidase E323A mutant bound to L-arginine | | 分子名称: | ARGININE, SODIUM ION, ZINC ION, ... | | 著者 | Agrawal, R, Kumar, A, Kumar, A, Makde, R.D. | | 登録日 | 2019-08-13 | | 公開日 | 2020-01-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis for the unusual substrate specificity of unique two-domain M1 metallopeptidase.
Int.J.Biol.Macromol., 147, 2020
|
|
6KOZ
 
 | | Crystal structure of two domain M1 zinc metallopeptidase E323 mutant bound to L-Leucine amino acid | | 分子名称: | LEUCINE, SODIUM ION, ZINC ION, ... | | 著者 | Agrawal, R, Kumar, A, Kumar, A, Makde, R.D. | | 登録日 | 2019-08-13 | | 公開日 | 2020-01-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural basis for the unusual substrate specificity of unique two-domain M1 metallopeptidase.
Int.J.Biol.Macromol., 147, 2020
|
|
6URI
 
 | |
5QTZ
 
 | | TGF-BETA RECEPTOR TYPE 1 KINASE DOMAIN (T204D) IN COMPLEX WITH 6-[1-(2,2-DIFLUOROETHYL)-4-(6-METHYLPYRIDIN-2-YL)-1H-IMIDAZOL-5-YL]IMIDAZO[1,2-A]PYRIDINE | | 分子名称: | 6-[1-(2,2-difluoroethyl)-4-(6-methylpyridin-2-yl)-1H-imidazol-5-yl]imidazo[1,2-a]pyridine, GLYCEROL, TGF-beta receptor type-1 | | 著者 | Sheriff, S. | | 登録日 | 2019-11-19 | | 公開日 | 2020-02-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Discovery of BMS-986260, a Potent, Selective, and Orally Bioavailable TGF beta R1 Inhibitor as an Immuno-oncology Agent.
Acs Med.Chem.Lett., 11, 2020
|
|
5QU0
 
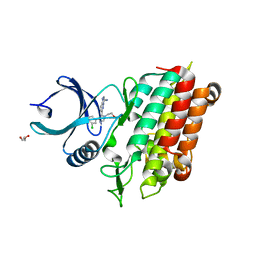 | | TGF-BETA RECEPTOR TYPE 1 KINASE DOMAIN (T204D) IN COMPLEX WITH 6-[4-(3-CHLORO-4-FLUOROPHENYL)-1-(2-HYDROXYETHYL)-1H-IMIDAZOL-5-YL]IMIDAZO[1,2-B]PYRIDAZINE-3-CARBONITRILE | | 分子名称: | 6-[4-(3-chloro-4-fluorophenyl)-1-(2-hydroxyethyl)-1H-imidazol-5-yl]imidazo[1,2-b]pyridazine-3-carbonitrile, GLYCEROL, TGF-beta receptor type-1 | | 著者 | Sheriff, S. | | 登録日 | 2019-11-19 | | 公開日 | 2020-02-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Discovery of BMS-986260, a Potent, Selective, and Orally Bioavailable TGF beta R1 Inhibitor as an Immuno-oncology Agent.
Acs Med.Chem.Lett., 11, 2020
|
|
5SW5
 
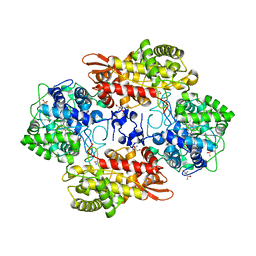 | | Crystal structure of native catalase-peroxidase KatG at pH7.5 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Catalase-peroxidase, ... | | 著者 | Loewen, P.C. | | 登録日 | 2016-08-08 | | 公開日 | 2016-08-24 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | A molecular switch and electronic circuit modulate catalase activity in catalase-peroxidases.
EMBO Rep., 6, 2005
|
|
5SX3
 
 | |
5SW6
 
 | |
5SX0
 
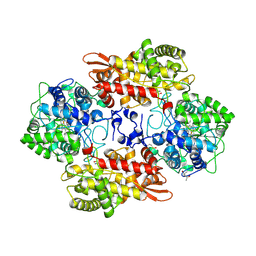 | |
5SX7
 
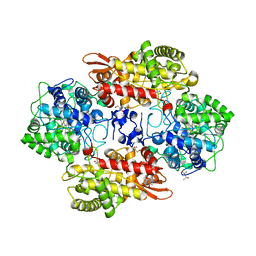 | | Crystal structure of catalase-peroxidase KatG of B. pseudomallei at pH 8.5 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Catalase-peroxidase, ... | | 著者 | Loewen, P.C. | | 登録日 | 2016-08-09 | | 公開日 | 2016-08-31 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Roles for Arg426 and Trp111 in the modulation of NADH oxidase activity of the catalase-peroxidase KatG from Burkholderia pseudomallei inferred from pH-induced structural changes.
Biochemistry, 45, 2006
|
|
5SX6
 
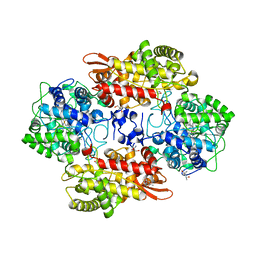 | | Crystal structure of the catalase-peroxidase KatG of B. pseudomallei at pH 6.5 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Catalase-peroxidase, ... | | 著者 | Loewen, P.C. | | 登録日 | 2016-08-09 | | 公開日 | 2016-08-31 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Roles for Arg426 and Trp111 in the modulation of NADH oxidase activity of the catalase-peroxidase KatG from Burkholderia pseudomallei inferred from pH-induced structural changes.
Biochemistry, 45, 2006
|
|
5SW4
 
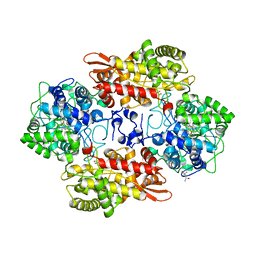 | |
8AVA
 
 | |
8AWH
 
 | |
5A7Z
 
 | | Crystal structure of Sulfolobus acidocaldarius Trm10 at 2.1 angstrom resolution. | | 分子名称: | TRNA (ADENINE(9)-N1)-METHYLTRANSFERASE | | 著者 | Van Laer, B, Roovers, M, Wauters, L, Kasprzak, J, Dyzma, M, Deyaert, E, Feller, A, Bujnicki, J, Droogmans, L, Versees, W. | | 登録日 | 2015-07-10 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural and Functional Insights Into tRNA Binding and Adenosine N1-Methylation by an Archaeal Trm10 Homologue.
Nucleic Acids Res., 44, 2016
|
|
