5E9G
 
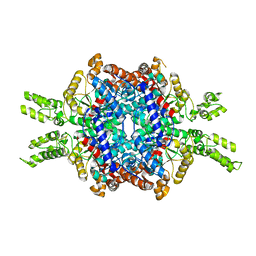 | | Structural insights of isocitrate lyases from Magnaporthe oryzae | | 分子名称: | GLYCEROL, GLYOXYLIC ACID, Isocitrate lyase, ... | | 著者 | Park, Y, Cho, Y, Lee, Y.-H, Lee, Y.-W, Rhee, S. | | 登録日 | 2015-10-15 | | 公開日 | 2016-04-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure and functional analysis of isocitrate lyases from Magnaporthe oryzae and Fusarium graminearum
J.Struct.Biol., 194, 2016
|
|
5E9H
 
 | | Structural insights of isocitrate lyases from Fusarium graminearum | | 分子名称: | Isocitrate lyase, MALONATE ION, MANGANESE (II) ION | | 著者 | Park, Y, Cho, Y, Lee, Y.-H, Lee, Y.-W, Rhee, S. | | 登録日 | 2015-10-15 | | 公開日 | 2016-04-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure and functional analysis of isocitrate lyases from Magnaporthe oryzae and Fusarium graminearum
J.Struct.Biol., 194, 2016
|
|
5E9F
 
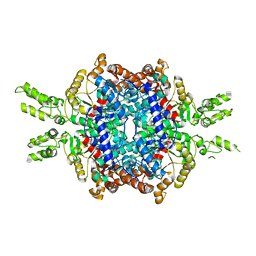 | | Structural insights of isocitrate lyases from Magnaporthe oryzae | | 分子名称: | Isocitrate lyase, MAGNESIUM ION | | 著者 | Park, Y, Cho, Y, Lee, Y.-H, Lee, Y.-W, Rhee, S. | | 登録日 | 2015-10-15 | | 公開日 | 2016-04-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure and functional analysis of isocitrate lyases from Magnaporthe oryzae and Fusarium graminearum
J.Struct.Biol., 194, 2016
|
|
7EF6
 
 | |
7EF7
 
 | |
4FFF
 
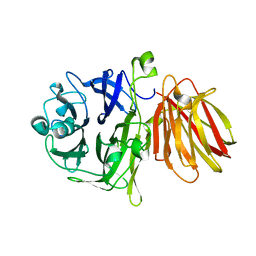 | |
4FFI
 
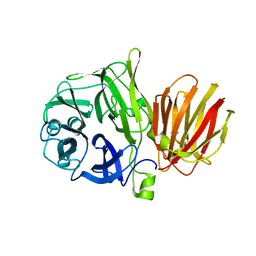 | |
5XU6
 
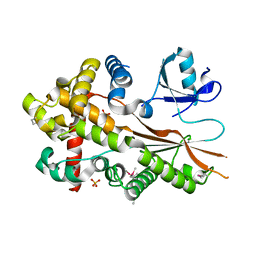 | | Crystal structure of inositol 1,3,4,5,6-pentakisphosphate 2-kinase (IPK1) from Cryptococcus neoformans | | 分子名称: | Inositol-pentakisphosphate 2-kinase, SULFATE ION | | 著者 | Oh, J, Rhee, S. | | 登録日 | 2017-06-22 | | 公開日 | 2017-10-04 | | 最終更新日 | 2017-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Crystal structure of inositol 1,3,4,5,6-pentakisphosphate 2-kinase from Cryptococcus neoformans.
J. Struct. Biol., 200, 2017
|
|
7VTE
 
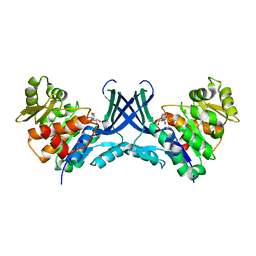 | |
7VTG
 
 | | Pseudouridine bound structure of Pseudouridine kinase (PUKI) S30A mutant from Escherichia coli strain B | | 分子名称: | 5-[(2~{S},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-1~{H}-pyrimidine-2,4-dione, Pseudouridine kinase | | 著者 | Kim, S.H, Rhee, S. | | 登録日 | 2021-10-29 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.89859128 Å) | | 主引用文献 | Substrate-binding loop interactions with pseudouridine trigger conformational changes that promote catalytic efficiency of pseudouridine kinase PUKI.
J.Biol.Chem., 298, 2022
|
|
7VTD
 
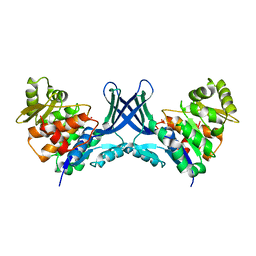 | |
7VTF
 
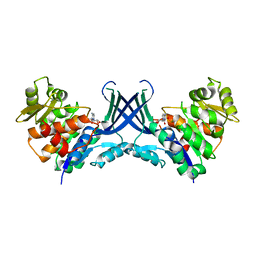 | |
7VVA
 
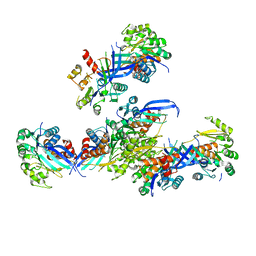 | | Pseudouridine bound structure of Pseudouridine kinase (PUKI) from Escherichia coli strain B | | 分子名称: | 5-[(2~{S},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-1~{H}-pyrimidine-2,4-dione, Pseudouridine kinase | | 著者 | Kim, S.H, Rhee, S. | | 登録日 | 2021-11-05 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.75029182 Å) | | 主引用文献 | Substrate-binding loop interactions with pseudouridine trigger conformational changes that promote catalytic efficiency of pseudouridine kinase PUKI.
J.Biol.Chem., 298, 2022
|
|
7VRX
 
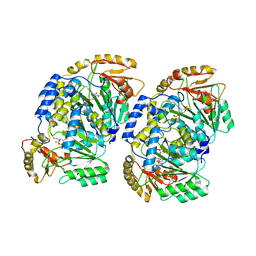 | | Pad-1 in the absence of substrate | | 分子名称: | Aminotransferase, SULFATE ION | | 著者 | Choi, M, Rhee, S. | | 登録日 | 2021-10-25 | | 公開日 | 2022-04-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.96634674 Å) | | 主引用文献 | Structural and biochemical basis for the substrate specificity of Pad-1, an indole-3-pyruvic acid aminotransferase in auxin homeostasis.
J.Struct.Biol., 214, 2022
|
|
3OUM
 
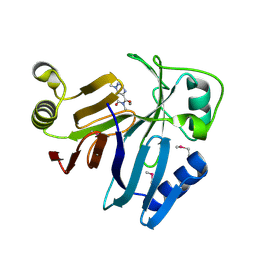 | | Crystal Structure of toxoflavin-degrading enzyme in complex with toxoflavin | | 分子名称: | 1,6-dimethylpyrimido[5,4-e][1,2,4]triazine-5,7(1H,6H)-dione, MANGANESE (II) ION, toxoflavin-degrading enzyme | | 著者 | Kim, M.I, Rhee, S. | | 登録日 | 2010-09-15 | | 公開日 | 2011-08-10 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and functional analysis of phytotoxin toxoflavin-degrading enzyme
Plos One, 6, 2011
|
|
3OUL
 
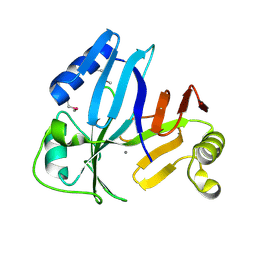 | |
4OK7
 
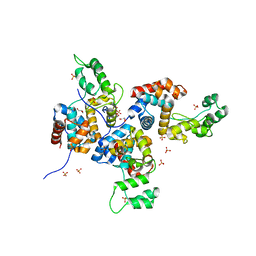 | | Structure of bacteriophage SPN1S endolysin from Salmonella typhimurium | | 分子名称: | Endolysin, GLYCEROL, SULFATE ION | | 著者 | Park, Y, Lim, J, Kong, M, Ryu, S, Rhee, S. | | 登録日 | 2014-01-22 | | 公開日 | 2014-03-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of bacteriophage SPN1S endolysin reveals an unusual two-module fold for the peptidoglycan lytic and binding activity.
Mol.Microbiol., 92, 2014
|
|
4P5F
 
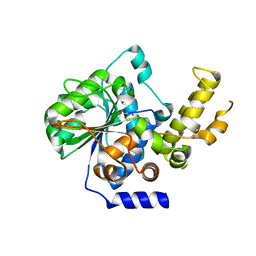 | | The crystal structure of type III effector protein XopQ complexed with adenosine diphosphate ribose | | 分子名称: | CALCIUM ION, Inosine-uridine nucleoside N-ribohydrolase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | 著者 | Yu, S, Hwang, I, Rhee, S. | | 登録日 | 2014-03-17 | | 公開日 | 2014-08-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The crystal structure of type III effector protein XopQ from Xanthomonas oryzae complexed with adenosine diphosphate ribose.
Proteins, 82, 2014
|
|
3HQ0
 
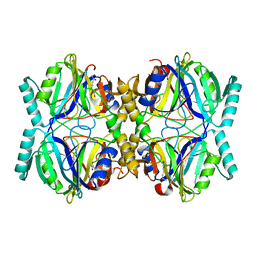 | | Crystal Structure Analysis of the 2,3-dioxygenase LapB from Pseudomonas in complex with a product | | 分子名称: | (2E,4E)-2-hydroxy-6-oxohepta-2,4-dienoic acid, Catechol 2,3-dioxygenase, FE (III) ION | | 著者 | Cho, J.-H, Rhee, S. | | 登録日 | 2009-06-05 | | 公開日 | 2009-10-20 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure and functional analysis of the extradiol dioxygenase LapB from a long-chain alkylphenol degradation pathway in Pseudomonas
J.Biol.Chem., 284, 2009
|
|
3HPV
 
 | |
3HPY
 
 | |
4E2Q
 
 | |
4E2S
 
 | |
3CZG
 
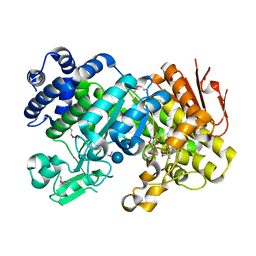 | |
3CZL
 
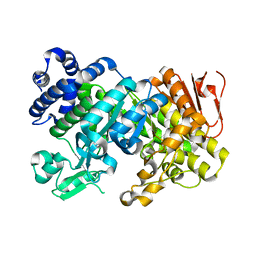 | |
