4B7W
 
 | | Ligand binding domain human hepatocyte nuclear factor 4alpha: Apo form | | 分子名称: | HEPATOCYTE NUCLEAR FACTOR 4-ALPHA | | 著者 | Dudasova, Z, Okvist, M, Kretova, M, Ondrovicova, G, Skrabana, R, LeGuevel, R, Salbert, G, Leonard, G, McSweeney, S, Barath, P. | | 登録日 | 2012-08-24 | | 公開日 | 2013-09-11 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (4 Å) | | 主引用文献 | Fatty Acids are not Essential Structural Components of Hepatocyte Nuclear Factor 4Alpha
To be Published
|
|
2V1C
 
 | |
2W4E
 
 | | Structure of an N-terminally truncated Nudix hydrolase DR2204 from Deinococcus radiodurans | | 分子名称: | MUTT/NUDIX FAMILY PROTEIN | | 著者 | Goncalves, A.M.D, Fioravanti, E, Stelter, M, McSweeney, S. | | 登録日 | 2008-11-25 | | 公開日 | 2009-12-01 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of an N-Terminally Truncated Nudix Hydrolase Dr2204 from Deinococcus Radiodurans.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
1V0U
 
 | |
1V0Y
 
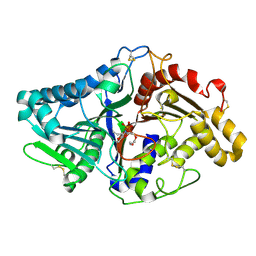 | |
7MHN
 
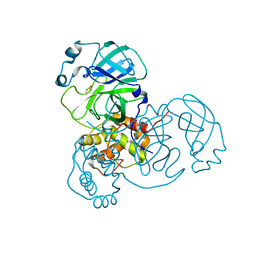 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 277 K | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | 著者 | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | 登録日 | 2021-04-15 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1908 Å) | | 主引用文献 | The temperature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro )
Iucrj, 9, 2022
|
|
1N7A
 
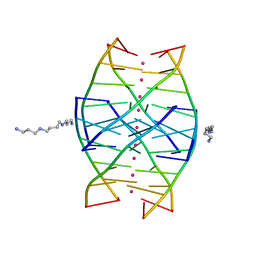 | | RIP-Radiation-damage Induced Phasing | | 分子名称: | POTASSIUM ION, RNA/DNA (5'-R(*U)-D(P*(BGM))-R(P*AP*GP*GP*U)-3'), SPERMINE | | 著者 | Ravelli, R.B.G, Leiros, H.-K.S, Pan, B, Caffrey, M, McSweeney, S. | | 登録日 | 2002-11-13 | | 公開日 | 2003-03-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Specific Radiation-Damage Can Be Used To Solve Macromolecular Crystal Structures
Structure, 11, 2003
|
|
7MHJ
 
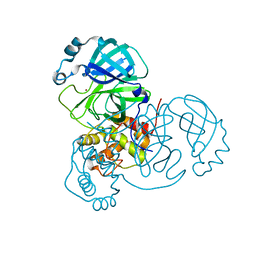 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 298 K and High Humidity | | 分子名称: | 3C-like proteinase, ZINC ION | | 著者 | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | 登録日 | 2021-04-15 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.0005 Å) | | 主引用文献 | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MHO
 
 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 298 K | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | 著者 | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | 登録日 | 2021-04-15 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | The temperature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro )
Iucrj, 9, 2022
|
|
7MHI
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 298 K | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | 著者 | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | 登録日 | 2021-04-15 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MHK
 
 | | Crystal Structure of Apo/Unliganded SARS-CoV-2 Main Protease (Mpro) at 310 K | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | 著者 | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | 登録日 | 2021-04-15 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9601 Å) | | 主引用文献 | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MNG
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor VBY-825 (Partial Occupancy) | | 分子名称: | (2R,3S)-N-cyclopropyl-3-{[(2R)-3-(cyclopropylmethanesulfonyl)-2-{[(1S)-2,2,2-trifluoro-1-(4-fluorophenyl)ethyl]amino}propanoyl]amino}-2-hydroxypentanamide (non-preferred name), 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Andi, B, Kumaran, D, Soares, A.S, Kreitler, D.F, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | 登録日 | 2021-04-30 | | 公開日 | 2021-05-12 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7MHQ
 
 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 310 K | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | 著者 | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | 登録日 | 2021-04-15 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9601 Å) | | 主引用文献 | The temperature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro )
Iucrj, 9, 2022
|
|
7MHP
 
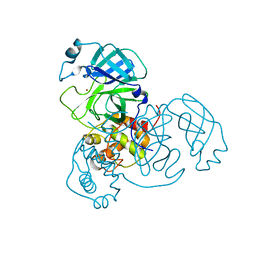 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 298 K at high humidity | | 分子名称: | 3C-like proteinase, ZINC ION | | 著者 | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | 登録日 | 2021-04-15 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.0005 Å) | | 主引用文献 | The temperature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro )
Iucrj, 9, 2022
|
|
1N7B
 
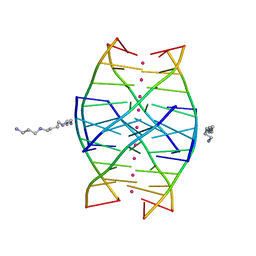 | | RIP-Radiation-damage Induced Phasing | | 分子名称: | POTASSIUM ION, RNA/DNA (5'-R(*U)-D(P*(BGM))-R(P*AP*GP*GP*U)-3'), SPERMINE | | 著者 | Ravelli, R.B.G, Leiros, H.-K.S, Pan, B, Caffrey, M, McSweeney, S. | | 登録日 | 2002-11-13 | | 公開日 | 2003-03-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Specific Radiation-Damage Can Be Used To Solve Macromolecular Crystal Structures
Structure, 11, 2003
|
|
7MHH
 
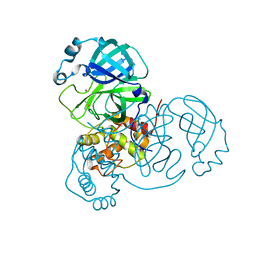 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 277 K | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | 著者 | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | 登録日 | 2021-04-15 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1908 Å) | | 主引用文献 | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MRR
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor Leupeptin | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, LEUPEPTIN | | 著者 | Andi, B, Kumaran, D, Soares, A.S, Kreitler, D.F, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | 登録日 | 2021-05-08 | | 公開日 | 2021-05-19 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
1N6X
 
 | | RIP-phasing on Bovine Trypsin | | 分子名称: | BENZYLAMINE, CALCIUM ION, GLYCEROL, ... | | 著者 | Ravelli, R.B.G, Leiros, H.-K.S, Pan, B, Caffrey, M, McSweeney, S. | | 登録日 | 2002-11-12 | | 公開日 | 2003-03-04 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Specific Radiation-Damage Can Be Used To Solve Macromolecular Crystal Structures
Structure, 11, 2003
|
|
1N6Y
 
 | | RIP-phasing on Bovine Trypsin | | 分子名称: | BENZYLAMINE, CALCIUM ION, GLYCEROL, ... | | 著者 | Ravelli, R.B.G, Leiros, H.-K.S, Pan, B, Caffrey, M, McSweeney, S. | | 登録日 | 2002-11-12 | | 公開日 | 2003-03-04 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Specific Radiation-Damage Can Be Used To Solve Macromolecular Crystal Structures
Structure, 11, 2003
|
|
1V0S
 
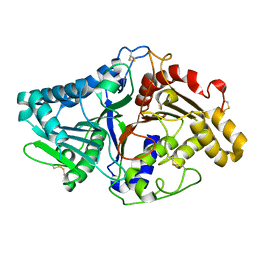 | |
1V0W
 
 | |
1V0T
 
 | |
1V0V
 
 | |
1V0R
 
 | |
2YG8
 
 | | Structure of an unusual 3-Methyladenine DNA Glycosylase II (Alka) from Deinococcus radiodurans | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, DNA-3-methyladenine glycosidase II, ... | | 著者 | Moe, E, Hall, D.R, Leiros, I, Talstad, V, Timmins, J, McSweeney, S. | | 登録日 | 2011-04-11 | | 公開日 | 2011-04-20 | | 最終更新日 | 2018-12-05 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-function studies of an unusual 3-methyladenine DNA glycosylase II (AlkA) from Deinococcus radiodurans.
Acta Crystallogr. D Biol. Crystallogr., 68, 2012
|
|
