5DT6
 
 | |
5DTB
 
 | |
5EHM
 
 | |
5EHS
 
 | |
3G3F
 
 | |
3G3H
 
 | |
8FNW
 
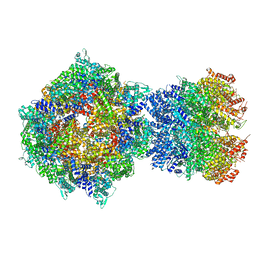 | | Structure of RdrA-RdrB complex from Escherichia coli RADAR defense system | | 分子名称: | Adenosine deaminase, Archaeal ATPase, ZINC ION | | 著者 | Duncan-Lowey, B, Johnson, A.G, Rawson, S, Mayer, M.L, Kranzusch, P.J. | | 登録日 | 2022-12-28 | | 公開日 | 2023-02-01 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (6.73 Å) | | 主引用文献 | Cryo-EM structure of the RADAR supramolecular anti-phage defense complex.
Cell, 186, 2023
|
|
8FNT
 
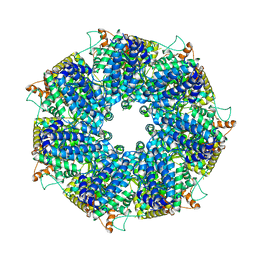 | | Structure of RdrA from Escherichia coli RADAR defense system | | 分子名称: | Archaeal ATPase | | 著者 | Duncan-Lowey, B, Johnson, A.G, Rawson, S, Mayer, M.L, Kranzusch, P.J. | | 登録日 | 2022-12-28 | | 公開日 | 2023-02-01 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.52 Å) | | 主引用文献 | Cryo-EM structure of the RADAR supramolecular anti-phage defense complex.
Cell, 186, 2023
|
|
8FNV
 
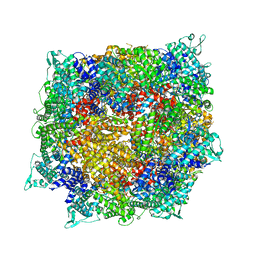 | | Structure of RdrB from Escherichia coli RADAR defense system | | 分子名称: | Adenosine deaminase, ZINC ION | | 著者 | Duncan-Lowey, B, Johnson, A.G, Rawson, S, Mayer, M.L, Kranzusch, P.J. | | 登録日 | 2022-12-28 | | 公開日 | 2023-02-01 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.11 Å) | | 主引用文献 | Cryo-EM structure of the RADAR supramolecular anti-phage defense complex.
Cell, 186, 2023
|
|
8FNU
 
 | | Structure of RdrA from Streptococcus suis RADAR defense system | | 分子名称: | KAP NTPase domain-containing protein | | 著者 | Duncan-Lowey, B, Johnson, A.G, Rawson, S, Mayer, M.L, Kranzusch, P.J. | | 登録日 | 2022-12-28 | | 公開日 | 2023-02-01 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Cryo-EM structure of the RADAR supramolecular anti-phage defense complex.
Cell, 186, 2023
|
|
8FDW
 
 | | Cryo-EM structure of SARS-CoV-2 postfusion spike in membrane | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S2, ... | | 著者 | Zhang, J, Shi, W, Cai, Y.F, Zhu, H.S, Peng, H.Q, Voyer, J, Volloch, S.R, Cao, H, Mayer, M.L, Song, K.K, Xu, C, Lu, J.M, Chen, B. | | 登録日 | 2022-12-05 | | 公開日 | 2023-05-10 | | 最終更新日 | 2023-07-26 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Cryo-EM structure of SARS-CoV-2 postfusion spike in membrane.
Nature, 619, 2023
|
|
3U93
 
 | |
3U92
 
 | |
3U94
 
 | |
3G3G
 
 | |
3G3K
 
 | |
3G3J
 
 | |
3H6H
 
 | |
3H6G
 
 | |
3G3I
 
 | |
1M5C
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH Br-HIBO AT 1.65 A RESOLUTION | | 分子名称: | (S)-2-AMINO-3-(4-BROMO-3-HYDROXY-ISOXAZOL-5-YL)PROPIONIC ACID, Glutamate receptor 2 | | 著者 | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | 登録日 | 2002-07-09 | | 公開日 | 2002-09-18 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
3OLZ
 
 | |
3OM1
 
 | |
3QLV
 
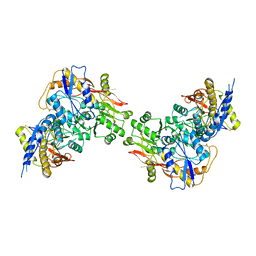 | |
3OM0
 
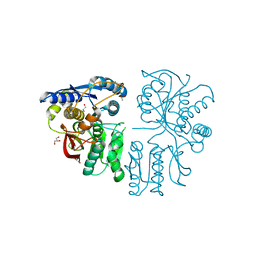 | |
