6XCJ
 
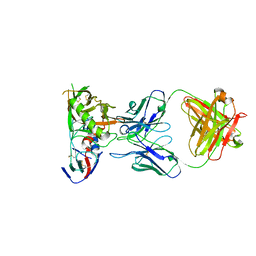 | | Crystal Structure of DH650 Fab from a Rhesus Macaque in Complex with HIV-1 gp120 Core | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DH650 Fab Heavy Chain, DH650 Fab Light Chain, ... | | 著者 | Raymond, D.D, Chug, H, Harrison, S.C. | | 登録日 | 2020-06-08 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Recapitulation of HIV-1 Env-antibody coevolution in macaques leading to neutralization breadth.
Science, 371, 2021
|
|
6WXF
 
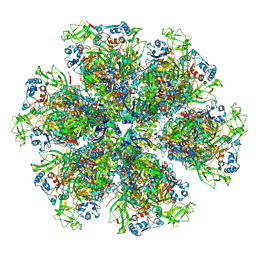 | | Cryo-EM reconstruction of VP5*/VP8* assembly from rhesus rotavirus particles - Intermediate conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Intermediate capsid protein VP6, ... | | 著者 | Herrmann, T, Harrison, S.C, Jenni, S. | | 登録日 | 2020-05-10 | | 公開日 | 2021-01-20 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Functional refolding of the penetration protein on a non-enveloped virus.
Nature, 590, 2021
|
|
6WXE
 
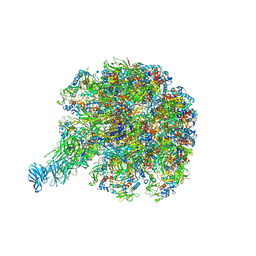 | | Cryo-EM reconstruction of VP5*/VP8* assembly from rhesus rotavirus particles - Upright conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Intermediate capsid protein VP6, ... | | 著者 | Herrmann, T, Harrison, S.C, Jenni, S. | | 登録日 | 2020-05-10 | | 公開日 | 2021-01-20 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Functional refolding of the penetration protein on a non-enveloped virus.
Nature, 590, 2021
|
|
6WUC
 
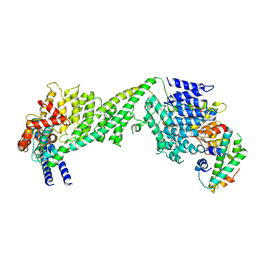 | | The yeast Ctf3 complex with Cnn1-Wip1 | | 分子名称: | Inner kinetochore subunit CNN1, Inner kinetochore subunit CTF3, Inner kinetochore subunit MCM16, ... | | 著者 | Hinshaw, S.M, Harrison, S.C. | | 登録日 | 2020-05-04 | | 公開日 | 2020-06-17 | | 最終更新日 | 2025-06-04 | | 実験手法 | ELECTRON MICROSCOPY (3.23 Å) | | 主引用文献 | The Structural Basis for Kinetochore Stabilization by Cnn1/CENP-T.
Curr.Biol., 30, 2020
|
|
5TD8
 
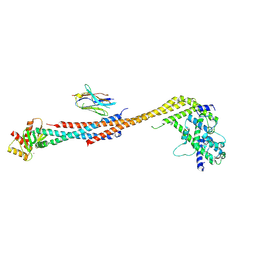 | | Crystal structure of an Extended Dwarf Ndc80 Complex | | 分子名称: | Kinetochore protein NDC80, Kinetochore protein NUF2, Kinetochore protein SPC24, ... | | 著者 | Valverde, R, Ingram, J, Harrison, S.C. | | 登録日 | 2016-09-17 | | 公開日 | 2016-11-16 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (7.531 Å) | | 主引用文献 | Conserved Tetramer Junction in the Kinetochore Ndc80 Complex.
Cell Rep, 17, 2016
|
|
5T6J
 
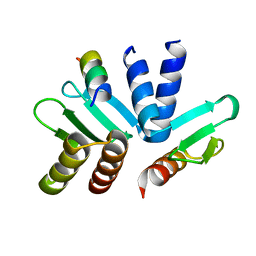 | | Structure of the MIND Complex Shows a Regulatory Focus of Yeast Kinetochore Assembly | | 分子名称: | Kinetochore protein SPC24, Kinetochore protein SPC25, Kinetochore-associated protein DSN1 | | 著者 | Valverde, R, Jenni, S, Dimitrova, Y, Khin, Y, Harrison, S.C. | | 登録日 | 2016-09-01 | | 公開日 | 2016-11-09 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.752 Å) | | 主引用文献 | Structure of the MIND Complex Defines a Regulatory Focus for Yeast Kinetochore Assembly.
Cell, 167, 2016
|
|
5T51
 
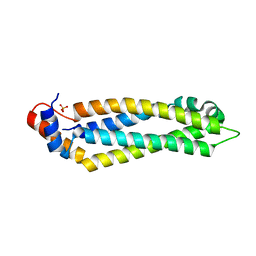 | | Structure of the MIND Complex Shows a Regulatory Focus of Yeast Kinetochore Assembly | | 分子名称: | KLLA0E05809p, KLLA0F02343p, SULFATE ION | | 著者 | Dimitrova, Y, Jenni, S, Valverde, R, Khin, Y, Harrison, S.C. | | 登録日 | 2016-08-30 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.2007 Å) | | 主引用文献 | Structure of the MIND Complex Defines a Regulatory Focus for Yeast Kinetochore Assembly.
Cell, 167, 2016
|
|
5T59
 
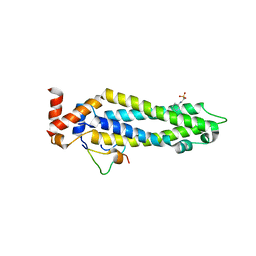 | | Structure of the MIND Complex Shows a Regulatory Focus of Yeast Kinetochore Assembly | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, KLLA0B13629p, KLLA0E05809p, ... | | 著者 | Dimitrova, Y, Jenni, S, Valverde, R, Khin, Y, Harrison, S.C. | | 登録日 | 2016-08-30 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.405 Å) | | 主引用文献 | Structure of the MIND Complex Defines a Regulatory Focus for Yeast Kinetochore Assembly.
Cell, 167, 2016
|
|
5TCS
 
 | |
5T58
 
 | | Structure of the MIND Complex Shows a Regulatory Focus of Yeast Kinetochore Assembly | | 分子名称: | KLLA0C15939p, KLLA0D15741p, KLLA0E05809p, ... | | 著者 | Dimitrova, Y, Jenni, S, Valverde, R, Khin, Y, Harrison, S.C. | | 登録日 | 2016-08-30 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.2131 Å) | | 主引用文献 | Structure of the MIND Complex Defines a Regulatory Focus for Yeast Kinetochore Assembly.
Cell, 167, 2016
|
|
5U0U
 
 | |
5U15
 
 | |
5TQA
 
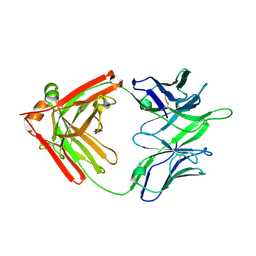 | |
5TRP
 
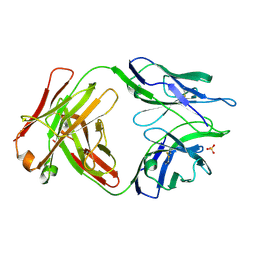 | |
5TPP
 
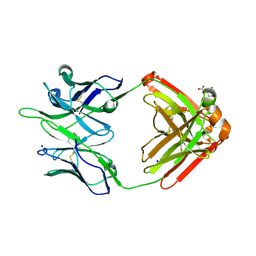 | |
5U0R
 
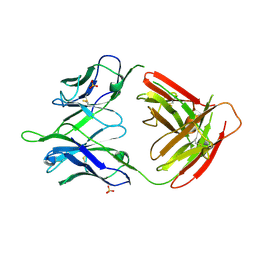 | |
5TPL
 
 | |
6U1X
 
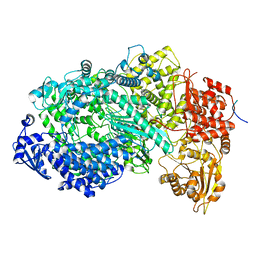 | | Structure of the Vesicular Stomatitis Virus L Protein in Complex with Its Phosphoprotein Cofactor (3.0 A resolution) | | 分子名称: | Phosphoprotein, RNA-directed RNA polymerase L, ZINC ION | | 著者 | Jenni, S, Bloyet, L.M, Dias-Avalos, R, Liang, B, Wheelman, S.P.J, Grigorieff, N, Harrison, S.C. | | 登録日 | 2019-08-17 | | 公開日 | 2020-01-22 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structure of the Vesicular Stomatitis Virus L Protein in Complex with Its Phosphoprotein Cofactor.
Cell Rep, 30, 2020
|
|
6UEB
 
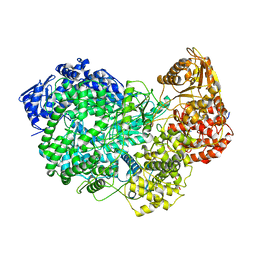 | |
6ULC
 
 | | Structure of full-length, fully glycosylated, non-modified HIV-1 gp160 bound to PG16 Fab at a nominal resolution of 4.6 Angstrom | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Pan, J, Chen, B, Harrison, S.C. | | 登録日 | 2019-10-07 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Cryo-EM Structure of Full-length HIV-1 Env Bound With the Fab of Antibody PG16.
J.Mol.Biol., 432, 2020
|
|
6UPH
 
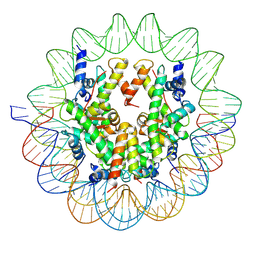 | | Structure of a Yeast Centromeric Nucleosome at 2.7 Angstrom resolution | | 分子名称: | DNA (119-MER), Histone H2A, Histone H2B.1, ... | | 著者 | Migl, D, Kschonsak, M, Arthur, C.P, Khin, Y, Harrison, S.C, Ciferri, C, Dimitrova, Y.N. | | 登録日 | 2019-10-17 | | 公開日 | 2019-11-06 | | 最終更新日 | 2025-05-14 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Cryoelectron Microscopy Structure of a Yeast Centromeric Nucleosome at 2.7 angstrom Resolution.
Structure, 28, 2020
|
|
6V05
 
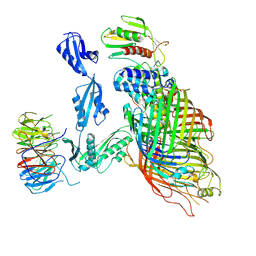 | | Cryo-EM structure of a substrate-engaged Bam complex | | 分子名称: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamA,Outer membrane protein assembly factor BamA,Outer membrane protein assembly factor BamA,Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | 著者 | Tomasek, D, Rawson, S, Lee, J, Wzorek, J.S, Harrison, S.C, Li, Z, Kahne, D. | | 登録日 | 2019-11-18 | | 公開日 | 2020-06-10 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structure of a nascent membrane protein as it folds on the BAM complex.
Nature, 583, 2020
|
|
4AU6
 
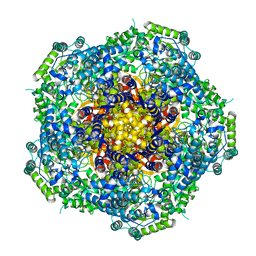 | | Location of the dsRNA-dependent polymerase, VP1, in rotavirus particles | | 分子名称: | RNA-DEPENDENT RNA POLYMERASE | | 著者 | Estrozi, L.F, Settembre, E.C, Goret, G, McClain, B, Zhang, X, Chen, J.Z, Grigorieff, N, Harrison, S.C. | | 登録日 | 2012-05-14 | | 公開日 | 2012-06-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (6 Å) | | 主引用文献 | Location of the Dsrna-Dependent Polymerase, Vp1, in Rotavirus Particles.
J.Mol.Biol., 425, 2013
|
|
3ZXA
 
 | |
3ZXU
 
 | |
