5KCM
 
 | | Crystal structure of iron-sulfur cluster containing photolyase PhrB mutant I51W | | 分子名称: | (6-4) photolyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | 著者 | Yang, X, Bowatte, K, Zhang, F, Lamparter, T. | | 登録日 | 2016-06-06 | | 公開日 | 2017-01-11 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.149 Å) | | 主引用文献 | Crystal Structures of Bacterial (6-4) Photolyase Mutants with Impaired DNA Repair Activity.
Photochem. Photobiol., 93, 2017
|
|
5LFA
 
 | | Crystal structure of iron-sulfur cluster containing bacterial (6-4) photolyase PhrB - Y424F mutant with impaired DNA repair activity | | 分子名称: | (6-4) photolyase, 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Kwiatkowski, D, Zhang, F, Krauss, N, Lamparter, T, Scheerer, P. | | 登録日 | 2016-06-30 | | 公開日 | 2017-01-11 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structures of Bacterial (6-4) Photolyase Mutants with Impaired DNA Repair Activity.
Photochem. Photobiol., 93, 2017
|
|
7TCV
 
 | | VDAC K12E mutant | | 分子名称: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Voltage-dependent anion-selective channel protein 1 | | 著者 | Khan, F, Abramson, J. | | 登録日 | 2021-12-28 | | 公開日 | 2023-01-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.604 Å) | | 主引用文献 | Dynamical control of the mitochondrial beta-barrel channel VDAC by electrostatic and mechanical coupling
To Be Published
|
|
7BX9
 
 | | Purification, characterization and X-ray structure of YhdA-type azoreductase from Bacillus velezensis | | 分子名称: | Azoreductase, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | 著者 | Khan, F, Suguna, K. | | 登録日 | 2020-04-18 | | 公開日 | 2020-12-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Purification, characterization, and crystal structure of YhdA-type azoreductase from Bacillus velezensis.
Proteins, 89, 2021
|
|
6DKL
 
 | | Crystal Structure of a Rationally Designed Six-Fold Symmetric DNA Scaffold | | 分子名称: | DNA (5'-D(P*CP*AP*CP*AP*CP*CP*GP*TP*AP*C)-3'), DNA (5'-D(P*GP*GP*AP*TP*GP*CP*AP*CP*A)-3'), DNA (5'-D(P*GP*TP*AP*CP*GP*GP*AP*TP*CP*CP*AP*G)-3'), ... | | 著者 | Simmons, C.R, Zhang, F, Yan, H. | | 登録日 | 2018-05-29 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.034 Å) | | 主引用文献 | Self-Assembly of a 3D DNA Crystal Structure with Rationally Designed Six-Fold Symmetry.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5T0Q
 
 | | Crystal structure of the Myc3 N-terminal domain [44-242] in complex with JAZ10 Jas domain [166-192] from arabidopsis | | 分子名称: | Protein TIFY 9, Transcription factor MYC3 | | 著者 | Ke, J, Zhang, F, Brunzelle, J.S, He, S.Y, Xu, H.E, Melcher, K. | | 登録日 | 2016-08-16 | | 公開日 | 2017-01-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structural insights into alternative splicing-mediated desensitization of jasmonate signaling.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5T0F
 
 | | Crystal structure of the Myc3 N-terminal domain [44-242] in complex with JAZ10 CMID domain [16-58] from arabidopsis | | 分子名称: | Protein TIFY 9, Transcription factor MYC3 | | 著者 | Ke, J, Zhang, F, Brunzelle, J.S, He, S.Y, Xu, H.E, Melcher, K. | | 登録日 | 2016-08-16 | | 公開日 | 2017-01-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural insights into alternative splicing-mediated desensitization of jasmonate signaling.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XXY
 
 | |
5UMO
 
 | |
5X1Y
 
 | |
6K07
 
 | | Crystal structure of REV7(R124A) in complex with a Shieldin3 fragment | | 分子名称: | Mitotic spindle assembly checkpoint protein MAD2B, SULFATE ION, Shieldin complex subunit 3 | | 著者 | Zhang, F, Dai, Y. | | 登録日 | 2019-05-05 | | 公開日 | 2019-12-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Structural basis for shieldin complex subunit 3-mediated recruitment of the checkpoint protein REV7 during DNA double-strand break repair.
J.Biol.Chem., 295, 2020
|
|
6K08
 
 | |
4IU6
 
 | | Human Methionine Aminopeptidase in complex with FZ1: Pyridinylquinazolines Selectively Inhibit Human Methionine Aminopeptidase-1 | | 分子名称: | 4-[4-(4-methoxyphenyl)piperazin-1-yl]-2-(pyridin-2-yl)quinazoline, COBALT (II) ION, Methionine aminopeptidase 1, ... | | 著者 | Gabelli, S.B, Zhang, F, Miller, M, Liu, J, Amzel, L.M. | | 登録日 | 2013-01-19 | | 公開日 | 2013-05-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Pyridinylquinazolines selectively inhibit human methionine aminopeptidase-1 in cells.
J.Med.Chem., 56, 2013
|
|
1RW8
 
 | | Crystal Structure of TGF-beta receptor I kinase with ATP site inhibitor | | 分子名称: | 3-(4-FLUOROPHENYL)-2-(6-METHYLPYRIDIN-2-YL)-5,6-DIHYDRO-4H-PYRROLO[1,2-B]PYRAZOLE, TGF-beta receptor type I | | 著者 | Zhang, F, Sawyer, J.S. | | 登録日 | 2003-12-16 | | 公開日 | 2005-02-01 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Synthesis and activity of new aryl- and heteroaryl-substituted 5,6-dihydro-4H-pyrrolo[1,2-b]pyrazole inhibitors of the transforming growth factor-beta type I receptor kinase domain.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1PYE
 
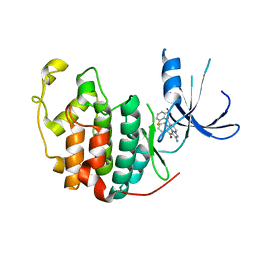 | | Crystal structure of CDK2 with inhibitor | | 分子名称: | Cell division protein kinase 2, [2-AMINO-6-(2,6-DIFLUORO-BENZOYL)-IMIDAZO[1,2-A]PYRIDIN-3-YL]-PHENYL-METHANONE | | 著者 | Zhang, F, Hamdouchi, C. | | 登録日 | 2003-07-08 | | 公開日 | 2004-07-13 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The discovery of a new structural class of cyclin-dependent kinase inhibitors, aminoimidazo[1,2-a]pyridines.
MOL.CANCER THER., 3, 2004
|
|
4DJA
 
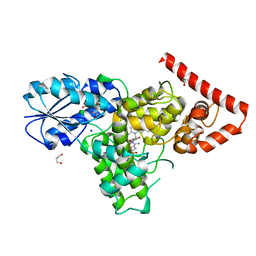 | | Crystal structure of a prokaryotic (6-4) photolyase PhrB from Agrobacterium Tumefaciens with an Fe-S cluster and a 6,7-dimethyl-8-ribityllumazine antenna chromophore at 1.45A resolution | | 分子名称: | 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | 著者 | Scheerer, P, Zhang, F, Oberpichler, I, Lamparter, T, Krauss, N. | | 登録日 | 2012-02-01 | | 公開日 | 2013-04-17 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Crystal structure of a prokaryotic (6-4) photolyase with an Fe-S cluster and a 6,7-dimethyl-8-ribityllumazine antenna chromophore.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1PY5
 
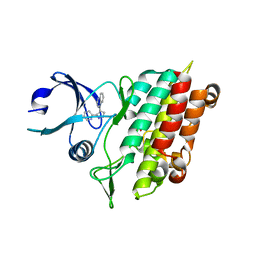 | | Crystal Structure of TGF-beta receptor I kinase with inhibitor | | 分子名称: | 4-(3-PYRIDIN-2-YL-1H-PYRAZOL-4-YL)QUINOLINE, SULFATE ION, TGF-beta receptor type I | | 著者 | Zhang, F, Sawyer, J.S. | | 登録日 | 2003-07-08 | | 公開日 | 2004-07-13 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Synthesis and activity of new aryl- and heteroaryl-substituted 5,6-dihydro-4H-pyrrolo[1,2-b]pyrazole inhibitors of the transforming growth factor-beta type I receptor kinase domain.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
4RQW
 
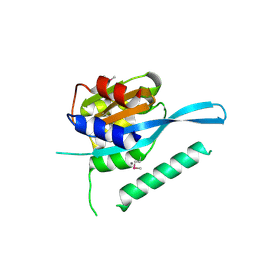 | | Crystal structure of Myc3 N-terminal JAZ-binding domain [44-238] from Arabidopsis | | 分子名称: | CALCIUM ION, Transcription factor MYC3 | | 著者 | Ke, J, Zhang, F, Zhou, X.E, Brunzelle, J, Zhou, M, Xu, H.E, Melcher, K, He, S.Y. | | 登録日 | 2014-11-05 | | 公開日 | 2015-08-12 | | 最終更新日 | 2015-09-16 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling.
Nature, 525, 2015
|
|
4RRU
 
 | | Myc3 N-terminal JAZ-binding domain[5-242] from arabidopsis | | 分子名称: | CALCIUM ION, Transcription factor MYC3 | | 著者 | Ke, J, Zhang, F, Zhou, X.E, Brunzelle, J.S, Zhou, M, Xu, H.E, Melcher, K, He, S.Y. | | 登録日 | 2014-11-06 | | 公開日 | 2015-08-12 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling.
Nature, 525, 2015
|
|
4RS9
 
 | | Structure of Myc3 N-terminal JAZ-binding domain [44-238] in complex with Jas motif of JAZ9 | | 分子名称: | Protein TIFY 7, Transcription factor MYC3 | | 著者 | Ke, J, Zhang, F, Zhou, X.E, Brunzelle, J.S, Zhou, M, Xu, H.E, Melcher, K, He, S.Y. | | 登録日 | 2014-11-07 | | 公開日 | 2015-08-12 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling.
Nature, 525, 2015
|
|
7ELV
 
 | |
7EXO
 
 | |
6IFB
 
 | |
6IFA
 
 | |
6LBP
 
 | | Structure of the Glutamine Phosphoribosylpyrophosphate Amidotransferase from Arabidopsis thaliana | | 分子名称: | Amidophosphoribosyltransferase 2, chloroplastic, IRON/SULFUR CLUSTER | | 著者 | Yi, Z, Cao, X, Han, F, Feng, Y. | | 登録日 | 2019-11-14 | | 公開日 | 2020-04-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.065 Å) | | 主引用文献 | Crystal Structure of the Chloroplastic Glutamine Phosphoribosylpyrophosphate Amidotransferase GPRAT2 FromArabidopsis thaliana.
Front Plant Sci, 11, 2020
|
|
