2B8F
 
 | |
2B0G
 
 | |
2B8G
 
 | |
1Z7T
 
 | |
1ML6
 
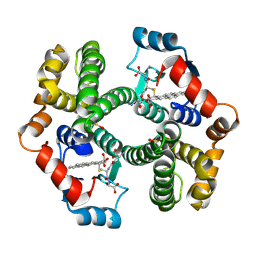 | | Crystal Structure of mGSTA2-2 in Complex with the Glutathione Conjugate of Benzo[a]pyrene-7(R),8(S)-Diol-9(S),10(R)-Epoxide | | 分子名称: | 2-AMINO-4-[1-(CARBOXYMETHYL-CARBAMOYL)-2-(9-HYDROXY-7,8-DIOXO-7,8,9,10-TETRAHYDRO-BENZO[DEF]CHRYSEN-10-YLSULFANYL)-ETHYLCARBAMOYL]-BUTYRIC ACID, Glutathione S-Transferase GT41A, ISOPROPYL ALCOHOL | | 著者 | Gu, Y, Xiao, B, Wargo, H.L, Bucher, M.H, Singh, S.V, Ji, X. | | 登録日 | 2002-08-30 | | 公開日 | 2003-04-15 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Residues 207, 216, and 221 and the catalytic activity of mGSTA1-1 and mGSTA2-2 toward
benzo[a]pyrene-(7R,8S)-diol-(9S,10R)-epoxide
Biochemistry, 42, 2003
|
|
2K4K
 
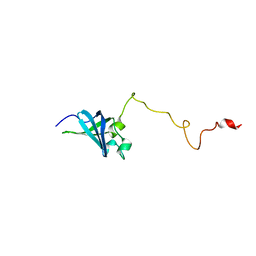 | | Solution structure of GSP13 from Bacillus subtilis | | 分子名称: | General stress protein 13 | | 著者 | Yu, W, Yu, B, Hu, J, Jin, C, Xia, B. | | 登録日 | 2008-06-13 | | 公開日 | 2009-05-12 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of GSP13 from Bacillus subtilis exhibits an S1 domain related to cold shock proteins.
J.Biomol.Nmr, 43, 2009
|
|
3IWM
 
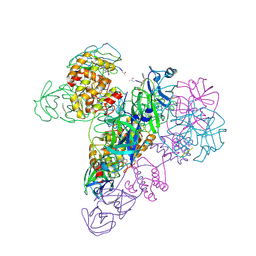 | | The octameric SARS-CoV main protease | | 分子名称: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | 著者 | Zhong, N, Zhang, S, Xue, F, Lou, Z, Rao, Z, Xia, B. | | 登録日 | 2009-09-02 | | 公開日 | 2010-07-21 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Three-dimensional domain swapping as a mechanism to lock the active conformation in a super-active octamer of SARS-CoV main protease
Protein Cell, 1, 2010
|
|
3EBN
 
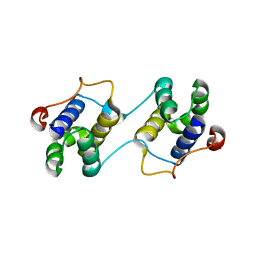 | | A Special Dimerization of SARS-CoV Main Protease C-Terminal Domain Due to Domain-swapping | | 分子名称: | Replicase polyprotein 1ab | | 著者 | Zhong, N, Zhang, S, Xue, F, Kang, X, Lou, Z, Xia, B. | | 登録日 | 2008-08-28 | | 公開日 | 2009-05-19 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | C-terminal domain of SARS-CoV main protease can form a 3D domain-swapped dimer
PROTEIN SCI., 18, 2009
|
|
2MXF
 
 | |
2MXE
 
 | |
5YTE
 
 | | Large fragment of DNA Polymerase I from Thermus aquaticus in a closed ternary complex with with natural dT:dATP base pair | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*TP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | 著者 | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Gao, Y.Q, Yi, C.Q. | | 登録日 | 2017-11-17 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
5YTD
 
 | | large fragment of DNA Polymerase I from Thermus aquaticus in a closed ternary complex with the natural base pair 5fC:dGTP | | 分子名称: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*(5FC)P*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | 著者 | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Gao, Y.Q, Yi, C.Q. | | 登録日 | 2017-11-17 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
5YTC
 
 | | Large fragment of DNA Polymerase I from Thermus aquaticus in a closed ternary complex with the unnatural base M-fC pair with dATP in the active site | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*(92F)P*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC)P*(DOC))-3'), ... | | 著者 | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Yi, C.Q. | | 登録日 | 2017-11-17 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
5YTF
 
 | | Structure of large fragment of DNA Polymerase I from Thermus aquaticus Host-Guest complex with the unnatural base M-fC pair with dA | | 分子名称: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*CP*GP*GP*CP*GP*CP*CP*GP*(92F)P*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | 著者 | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Gao, Y.Q, Yi, C.Q. | | 登録日 | 2017-11-17 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
5YTG
 
 | | Structure of large fragment of DNA Polymerase I from Thermus aquaticus Host-Guest complex with the unnatural base I-fC pair with dA | | 分子名称: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*CP*GP*GP*CP*GP*CP*CP*GP*(94O)P*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | 著者 | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Gao, Y.Q, Yi, C.Q. | | 登録日 | 2017-11-17 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
5YTH
 
 | | Structure of large fragment of DNA Polymerase I from Thermus aquaticus Host-Guest complex with the unnatural base M-fC pair with dG | | 分子名称: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*CP*GP*GP*CP*GP*CP*CP*GP*(92F)P*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*GP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | 著者 | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Gao, Y.Q, Yi, C.Q. | | 登録日 | 2017-11-17 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.53 Å) | | 主引用文献 | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
5Z3N
 
 | | Structure of large fragment of DNA Polymerase I from Thermus aquaticus Host-Guest complex with the unnatural base 5fC pair with dA | | 分子名称: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*CP*GP*GP*CP*GP*CP*CP*GP*(5FC)P*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | 著者 | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Gao, Y.Q, Yi, C.Q. | | 登録日 | 2018-01-08 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
2OZX
 
 | |
2OZW
 
 | |
2M5H
 
 | |
1J7C
 
 | | STRUCTURE OF THE ANABAENA FERREDOXIN MUTANT E95K | | 分子名称: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN I | | 著者 | Hurley, J.K, Weber-Main, A.M, Stankovich, M.T, Benning, M.M, Thoden, J.B, Vanhooke, J.L, Holden, H.M, Chae, Y.K, Xia, B, Cheng, H, Markley, J.L, Martinez-Julvez, M, Gomez-Moreno, C, Schmeits, J.L, Tollin, G. | | 登録日 | 2001-05-16 | | 公開日 | 2001-05-23 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure-function relationships in Anabaena ferredoxin: correlations between X-ray crystal structures, reduction potentials, and rate constants of electron transfer to ferredoxin:NADP+ reductase for site-specific ferredoxin mutants.
Biochemistry, 36, 1997
|
|
2MY9
 
 | |
5XF0
 
 | |
2HF6
 
 | | Solution structure of human zeta-COP | | 分子名称: | Coatomer subunit zeta-1 | | 著者 | Yu, W, Jin, C, Xia, B. | | 登録日 | 2006-06-23 | | 公開日 | 2007-06-12 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of human zeta-COP: direct evidences for structural similarity between COP I and clathrin-adaptor coats
J.Mol.Biol., 386, 2009
|
|
2KNG
 
 | | Solution structure of C-domain of Lsr2 | | 分子名称: | Protein lsr2 | | 著者 | Li, Y, Xia, B. | | 登録日 | 2009-08-22 | | 公開日 | 2010-03-16 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Lsr2 is a nucleoid-associated protein that targets AT-rich sequences and virulence genes in Mycobacterium tuberculosis
Proc.Natl.Acad.Sci.USA, 2010
|
|
