8GST
 
 | | Crystal structure of L-2,4-diketo-3-deoxyrhamnonate hydrolase from Sphingomonas sp. (pyruvate bound-form) | | 分子名称: | L-2,4-diketo-3-deoxyrhamnonate hydrolase, MAGNESIUM ION, PYRUVIC ACID | | 著者 | Fukuhara, S, Watanabe, Y, Watanabe, S, Nishiwaki, H. | | 登録日 | 2022-09-07 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Crystal Structure of l-2,4-Diketo-3-deoxyrhamnonate Hydrolase Involved in the Nonphosphorylated l-Rhamnose Pathway from Bacteria.
Biochemistry, 62, 2023
|
|
8GSR
 
 | | Crystal structure of L-2,4-diketo-3-deoxyrhamnonate hydrolase from Sphingomonas sp. (apo-form) | | 分子名称: | L-2,4-diketo-3-deoxyrhamnonate hydrolase, MAGNESIUM ION | | 著者 | Fukuhara, S, Watanabe, Y, Watanabe, S, Nishiwaki, H. | | 登録日 | 2022-09-07 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Crystal Structure of l-2,4-Diketo-3-deoxyrhamnonate Hydrolase Involved in the Nonphosphorylated l-Rhamnose Pathway from Bacteria.
Biochemistry, 62, 2023
|
|
7B81
 
 | |
7BYW
 
 | |
7BYU
 
 | | Crystal structure of Acidovorax avenae L-fucose mutarotase (apo form) | | 分子名称: | 1,2-ETHANEDIOL, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, L-fucose mutarotase | | 著者 | Watanabe, Y, Fukui, Y, Watanabe, S. | | 登録日 | 2020-04-24 | | 公開日 | 2020-05-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.206 Å) | | 主引用文献 | Functional and structural characterization of a novel L-fucose mutarotase involved in non-phosphorylative pathway of L-fucose metabolism.
Biochem.Biophys.Res.Commun., 528, 2020
|
|
1K2G
 
 | | Structural basis for the 3'-terminal guanosine recognition by the group I intron | | 分子名称: | 5'-R(*CP*AP*GP*AP*CP*UP*UP*CP*GP*GP*UP*CP*GP*CP*AP*GP*AP*GP*AP*UP*GP*G)-3' | | 著者 | Kitamura, Y, Muto, Y, Watanabe, S, Kim, I, Ito, T, Nishiya, Y, Sakamoto, K, Ohtsuki, T, Kawai, G, Watanabe, K, Hosono, K, Takaku, H, Katoh, E, Yamazaki, T, Inoue, T, Yokoyama, S. | | 登録日 | 2001-09-27 | | 公開日 | 2002-05-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of an RNA fragment with the P7/P9.0 region and the 3'-terminal guanosine of the tetrahymena group I intron.
RNA, 8, 2002
|
|
2LC2
 
 | | Solution structure of the RXLR effector P. capsici AVR3a4 | | 分子名称: | AVR3a4 | | 著者 | Li, H, Koshiba, S, Yaeno, T, Sato, M, Watanabe, S, Harada, T, Shirasu, K, Kigawa, T. | | 登録日 | 2011-04-12 | | 公開日 | 2011-08-03 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A PIP-binding interface in the oomycete RXLR effector AVR3A is required for its accumulation in host cells to modulate plant immunity
To be Published
|
|
6UD0
 
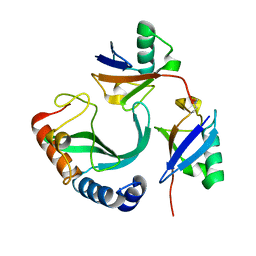 | | Solution-state NMR structural ensemble of human Tsg101 UEV in complex with K63-linked diubiquitin | | 分子名称: | Tumor susceptibility gene 101 protein, Ubiquitin | | 著者 | Strickland, M, Watanabe, S, Bonn, S.M, Camara, C.M, Fushman, D, Carter, C.A, Tjandra, N. | | 登録日 | 2019-09-18 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Tsg101/ESCRT-I Recruitment Regulated by the Dual Binding Modes of K63-Linked Diubiquitin
Structure, 2021
|
|
2KUQ
 
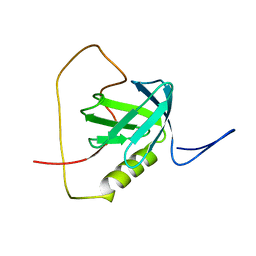 | | Solution structure of the chimera of the PTB domain of SNT-2 and 19-residue peptide (aa 1571-1589) of HALK | | 分子名称: | Fibroblast growth factor receptor substrate 3,LINKER,ALK tyrosine kinase receptor | | 著者 | Li, H, Koshiba, S, Tomizawa, T, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2010-02-24 | | 公開日 | 2010-05-26 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for the recognition of nucleophosmin-anaplastic lymphoma kinase oncoprotein by the phosphotyrosine binding domain of Suc1-associated neurotrophic factor-induced tyrosine-phosphorylated target-2
J.Struct.Funct.Genom., 11, 2010
|
|
4G9I
 
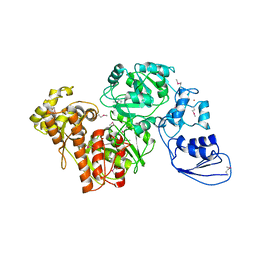 | | Crystal structure of T.kodakarensis HypF | | 分子名称: | Hydrogenase maturation protein HypF, ZINC ION | | 著者 | Tominaga, T, Watanabe, S, Matsumi, R, Atomi, H, Imanaka, T, Miki, K. | | 登録日 | 2012-07-24 | | 公開日 | 2012-10-24 | | 実験手法 | X-RAY DIFFRACTION (4.5 Å) | | 主引用文献 | Structure of the [NiFe]-hydrogenase maturation protein HypF from Thermococcus kodakarensis KOD1.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
6J7C
 
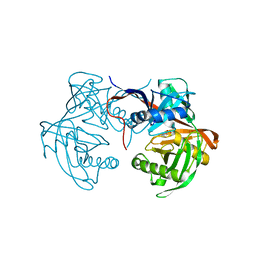 | | Crystal structure of proline racemase-like protein from Thermococcus litoralis in complex with proline | | 分子名称: | PROLINE, Proline racemase | | 著者 | Watanabe, Y, Watanabe, S, Itoh, Y, Watanabe, Y. | | 登録日 | 2019-01-17 | | 公開日 | 2019-02-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of substrate-bound bifunctional proline racemase/hydroxyproline epimerase from a hyperthermophilic archaeon.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
2LEX
 
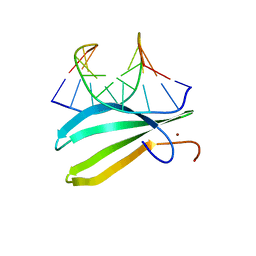 | | Complex of the C-terminal WRKY domain of AtWRKY4 and a W-box DNA | | 分子名称: | DNA (5'-D(*CP*G*CP*CP*TP*TP*TP*GP*AP*CP*CP*AP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*C*TP*GP*GP*TP*CP*AP*AP*AP*GP*GP*CP*G)-3'), Probable WRKY transcription factor 4, ... | | 著者 | Yamasaki, K, Kigawa, T, Watanabe, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2011-06-24 | | 公開日 | 2012-01-18 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for sequence-spscific DNA recognition by an Arabidopsis WRKY transcription factor
J.Biol.Chem., 2012
|
|
6JNJ
 
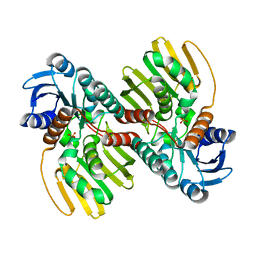 | |
6JNK
 
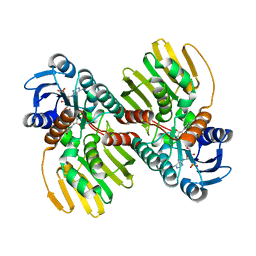 | |
1J1W
 
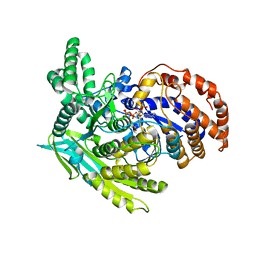 | | Crystal Structure Of The Monomeric Isocitrate Dehydrogenase In Complex With NADP+ | | 分子名称: | Isocitrate Dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Yasutake, Y, Watanabe, S, Yao, M, Takada, Y, Fukunaga, N, Tanaka, I. | | 登録日 | 2002-12-19 | | 公開日 | 2003-09-23 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Crystal Structure of the Monomeric Isocitrate Dehydrogenase in the Presence of NADP+
J.Biol.Chem., 278, 2003
|
|
1ITW
 
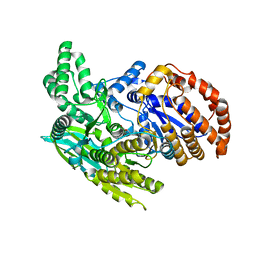 | | Crystal structure of the monomeric isocitrate dehydrogenase in complex with isocitrate and Mn | | 分子名称: | ISOCITRIC ACID, Isocitrate dehydrogenase, MANGANESE (II) ION | | 著者 | Yasutake, Y, Watanabe, S, Yao, M, Takada, Y, Fukunaga, N, Tanaka, I. | | 登録日 | 2002-02-12 | | 公開日 | 2002-12-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure of the Monomeric Isocitrate Dehydrogenase: Evidence of a Protein Monomerization by a Domain Duplication
Structure, 10, 2002
|
|
7Y9P
 
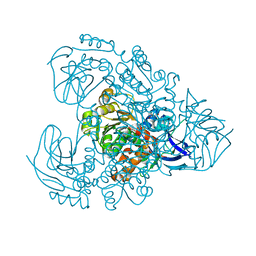 | | Xylitol dehydrogenase S96C/S99C/Y102C mutant(thermostabilized form) from Pichia stipitis | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | 著者 | Yoshiwara, K, Watanabe, Y, Watanabe, S. | | 登録日 | 2022-06-25 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Molecular evolutionary insight of structural zinc atom in yeast xylitol dehydrogenases and its application in bioethanol production by lignocellulosic biomass.
Sci Rep, 13, 2023
|
|
8J7W
 
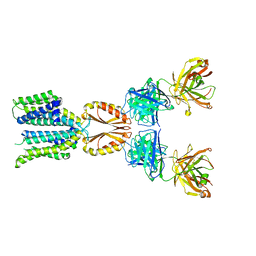 | | Cryo-EM structure of hZnT7-Fab complex in zinc state 2, determined in heterogeneous conformations- one subunit in an inward-facing zinc-bound and the other in an outward-facing zinc-bound conformation | | 分子名称: | Heavy chain of YN7114-08 Fab, Light chain of YN7114-08 Fab, ZINC ION, ... | | 著者 | Han, B.B, Inaba, K, Watanabe, S. | | 登録日 | 2023-04-28 | | 公開日 | 2023-09-20 | | 実験手法 | ELECTRON MICROSCOPY (2.92 Å) | | 主引用文献 | Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn 2+ uptake into the Golgi apparatus.
Nat Commun, 14, 2023
|
|
8J7X
 
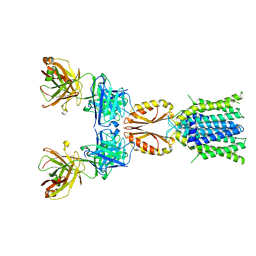 | | Cryo-EM structure of hZnT7DeltaHis-loop-Fab complex in zinc-unbound state, determined in outward-facing conformation | | 分子名称: | Heavy chain of YN7114-08 Fab, Light chain of YN7114-08 Fab, Zinc transporter 7 | | 著者 | Han, B.B, Inaba, K, Watanabe, S. | | 登録日 | 2023-04-28 | | 公開日 | 2023-09-20 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn 2+ uptake into the Golgi apparatus.
Nat Commun, 14, 2023
|
|
8J7V
 
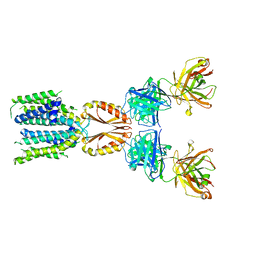 | | Cryo-EM structure of hZnT7-Fab complex in zinc-unbound state, determined in heterogeneous conformations- one subunit in an inward-facing and the other in an outward-facing conformation | | 分子名称: | Heavy chain of YN7114-08 Fab, Light chain of YN7114-08 Fab, Zinc transporter 7 | | 著者 | Han, B.B, Inaba, K, Watanabe, S. | | 登録日 | 2023-04-28 | | 公開日 | 2023-09-20 | | 実験手法 | ELECTRON MICROSCOPY (2.79 Å) | | 主引用文献 | Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn 2+ uptake into the Golgi apparatus.
Nat Commun, 14, 2023
|
|
8J7U
 
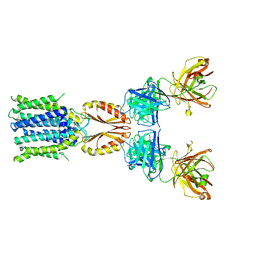 | | Cryo-EM structure of hZnT7-Fab complex in zinc-bound state, determined in outward-facing conformation | | 分子名称: | Heavy chain of YN7114-08 Fab, Light chain of YN7114-08 Fab, ZINC ION, ... | | 著者 | Han, B.B, Inaba, K, Watanabe, S. | | 登録日 | 2023-04-28 | | 公開日 | 2023-09-20 | | 実験手法 | ELECTRON MICROSCOPY (3.12 Å) | | 主引用文献 | Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn 2+ uptake into the Golgi apparatus.
Nat Commun, 14, 2023
|
|
8J80
 
 | | Cryo-EM structure of hZnT7-Fab complex in zinc state 1, determined in heterogeneous conformations- one subunit in an inward-facing zinc-bound and the other in an outward-facing zinc-unbound conformation | | 分子名称: | Heavy chain of YN7114-08 Fab, Light chain of YN7114-08 Fab, ZINC ION, ... | | 著者 | Han, B.B, Inaba, K, Watanabe, S. | | 登録日 | 2023-04-28 | | 公開日 | 2023-09-20 | | 実験手法 | ELECTRON MICROSCOPY (2.68 Å) | | 主引用文献 | Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn 2+ uptake into the Golgi apparatus.
Nat Commun, 14, 2023
|
|
8J7T
 
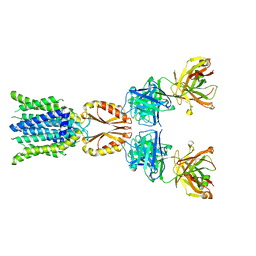 | | Cryo-EM structure of hZnT7-Fab complex in zinc-unbound state, determined in outward-facing conformation | | 分子名称: | Heavy chain of YN7114-08 Fab, Light chain of YN7114-08 Fab, Zinc transporter 7 | | 著者 | Han, B.B, Inaba, K, Watanabe, S. | | 登録日 | 2023-04-28 | | 公開日 | 2023-09-20 | | 実験手法 | ELECTRON MICROSCOPY (2.2 Å) | | 主引用文献 | Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn 2+ uptake into the Golgi apparatus.
Nat Commun, 14, 2023
|
|
8J7Y
 
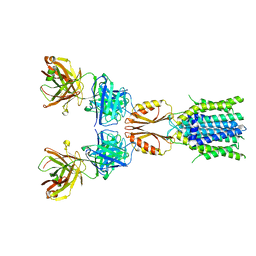 | | Cryo-EM structure of hZnT7DeltaHis-loop-Fab complex in zinc-bound state, determined in outward-facing conformation | | 分子名称: | Heavy chain of YN7114-08 Fab, Light chain of YN7114-08 Fab, ZINC ION, ... | | 著者 | Han, B.B, Inaba, K, Watanabe, S. | | 登録日 | 2023-04-28 | | 公開日 | 2023-09-20 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn 2+ uptake into the Golgi apparatus.
Nat Commun, 14, 2023
|
|
6L06
 
 | |
