6AEE
 
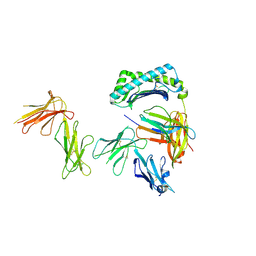 | | Crystal structure of the four Ig-like domains of LILRB1 complexed with HLA-G | | 分子名称: | 9 Mer Peptide (RL9) From Histone H2A.x, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | 著者 | Wang, Q, Song, H, Qi, J, Gao, G.F. | | 登録日 | 2018-08-04 | | 公開日 | 2019-07-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.303 Å) | | 主引用文献 | Structures of the four Ig-like domain LILRB2 and the four-domain LILRB1 and HLA-G1 complex.
Cell. Mol. Immunol., 2019
|
|
6AED
 
 | |
3SVO
 
 | |
3SVS
 
 | |
3SVN
 
 | |
3SVU
 
 | |
3SVR
 
 | |
5ZY5
 
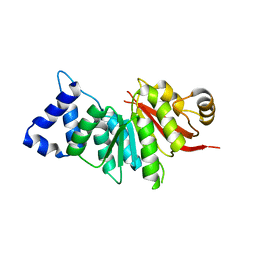 | | spCOMT apo structure | | 分子名称: | Probable catechol O-methyltransferase 1 | | 著者 | Wang, Q, Xu, L. | | 登録日 | 2018-05-22 | | 公開日 | 2019-05-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.295 Å) | | 主引用文献 | structural and functional investigations of spCOMT
To Be Published
|
|
5ZY6
 
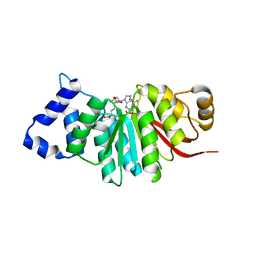 | | catechol methyltransferase spCOMT | | 分子名称: | Probable catechol O-methyltransferase 1, S-ADENOSYLMETHIONINE | | 著者 | Wang, Q, Xu, L. | | 登録日 | 2018-05-22 | | 公開日 | 2019-05-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.099 Å) | | 主引用文献 | structural insight into spCOMT
To Be Published
|
|
7C2K
 
 | | COVID-19 RNA-dependent RNA polymerase pre-translocated catalytic complex | | 分子名称: | Non-structural protein 7, Non-structural protein 8, RNA (29-MER), ... | | 著者 | Wang, Q, Gao, Y, Ji, W, Mu, A, Rao, Z. | | 登録日 | 2020-05-07 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.93 Å) | | 主引用文献 | Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase.
Cell, 182, 2020
|
|
6K87
 
 | |
6K82
 
 | | RGLG1 mutant-D338A E378A | | 分子名称: | E3 ubiquitin-protein ligase RGLG1, MAGNESIUM ION, SODIUM ION | | 著者 | Wang, Q, Wu, Y. | | 登録日 | 2019-06-11 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.402 Å) | | 主引用文献 | RGLG1 mutant-D338A E378A
To Be Published
|
|
6K89
 
 | | The closed state of RGLG1 VWA domain | | 分子名称: | E3 ubiquitin-protein ligase RGLG1, MAGNESIUM ION, SODIUM ION | | 著者 | Wang, Q, Wu, Y. | | 登録日 | 2019-06-11 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.689 Å) | | 主引用文献 | The closed state of RGLG1 VWA domain
To Be Published
|
|
6K8E
 
 | | Global regulatory element SarX | | 分子名称: | HTH-type transcriptional regulator SarX | | 著者 | Wang, Q. | | 登録日 | 2019-06-11 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.002 Å) | | 主引用文献 | Crystal structure of SarX from Staphylococcus aureus
To Be Published
|
|
6K85
 
 | |
6K8A
 
 | |
6K88
 
 | | RGLG1 MIDAS binds calcium ion | | 分子名称: | CALCIUM ION, E3 ubiquitin-protein ligase RGLG1 | | 著者 | Wang, Q, Wu, Y. | | 登録日 | 2019-06-11 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.788 Å) | | 主引用文献 | RGLG1 MIDAS binds calcium ion
To Be Published
|
|
5XYB
 
 | |
5Y24
 
 | | Crystal structure of AimR from Bacillus phage SPbeta in complex with its signalling peptide | | 分子名称: | AimR transcriptional regulator, BROMIDE ION, GLY-MET-PRO-ARG-GLY-ALA | | 著者 | Wang, Q, Guan, Z.Y, Zou, T.T, Yin, P. | | 登録日 | 2017-07-24 | | 公開日 | 2018-09-19 | | 最終更新日 | 2018-11-28 | | 実験手法 | X-RAY DIFFRACTION (1.922 Å) | | 主引用文献 | Structural basis of the arbitrium peptide-AimR communication system in the phage lysis-lysogeny decision.
Nat Microbiol, 3, 2018
|
|
8T3P
 
 | |
8W5K
 
 | | Cryo-EM structure of the yeast TOM core complex crosslinked by BS3 (from TOM-TIM23 complex) | | 分子名称: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | 著者 | Wang, Q, Guan, Z.Y, Zhuang, J.J, Huang, R, Yin, P. | | 登録日 | 2023-08-27 | | 公開日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | The architecture of substrate-engaged TOM-TIM23 supercomplex reveals preprotein proximity sites for mitochondrial protein translocation.
Cell Discov, 10, 2024
|
|
8W5J
 
 | | Cryo-EM structure of the yeast TOM core complex (from TOM-TIM23 complex) | | 分子名称: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | 著者 | Wang, Q, Guan, Z.Y, Zhuang, J.J, Huang, R, Yin, P. | | 登録日 | 2023-08-27 | | 公開日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | The architecture of substrate-engaged TOM-TIM23 supercomplex reveals preprotein proximity sites for mitochondrial protein translocation.
Cell Discov, 10, 2024
|
|
5YGU
 
 | | Crystal structure of Escherichia coli (strain K12) mRNA Decapping Complex RppH-DapF | | 分子名称: | Diaminopimelate epimerase, IODIDE ION, L(+)-TARTARIC ACID, ... | | 著者 | Wang, Q, Guan, Z.Y, Zhang, D.L, Zou, T.T, Yin, P. | | 登録日 | 2017-09-27 | | 公開日 | 2018-06-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.298 Å) | | 主引用文献 | DapF stabilizes the substrate-favoring conformation of RppH to stimulate its RNA-pyrophosphohydrolase activity in Escherichia coli.
Nucleic Acids Res., 46, 2018
|
|
5YWJ
 
 | | Global regulatory element SarX | | 分子名称: | HTH-type transcriptional regulator SarX | | 著者 | Wang, Q. | | 登録日 | 2017-11-29 | | 公開日 | 2018-12-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.102 Å) | | 主引用文献 | Crystal structure of SarX from Staphylococcus aureus
To Be Published
|
|
8HMG
 
 | | The open state of RGLG2-VWA | | 分子名称: | CALCIUM ION, E3 ubiquitin-protein ligase RGLG2, MAGNESIUM ION, ... | | 著者 | Wang, Q. | | 登録日 | 2022-12-03 | | 公開日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Ca2+-based Allosteric Switches and Shape Shifting in RGLG1 VWA domain
To Be Published
|
|
