4XBU
 
 | | In vitro Crystal Structure of PAK4 in complex with Inka peptide | | 分子名称: | Protein FAM212A, Serine/threonine-protein kinase PAK 4 | | 著者 | Baskaran, Y, Ang, K.C, Anekal, P.V, Chan, W.L, Grimes, J.M, Manser, E, Robinson, R.C. | | 登録日 | 2014-12-17 | | 公開日 | 2015-12-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | An in cellulo-derived structure of PAK4 in complex with its inhibitor Inka1
Nat Commun, 6, 2015
|
|
4XGC
 
 | | Crystal structure of the eukaryotic origin recognition complex | | 分子名称: | CHLORIDE ION, Origin recognition complex subunit 1, Origin recognition complex subunit 2, ... | | 著者 | Bleichert, F, Botchan, M.R, Berger, J.M. | | 登録日 | 2014-12-30 | | 公開日 | 2015-04-01 | | 最終更新日 | 2019-12-04 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Crystal structure of the eukaryotic origin recognition complex.
Nature, 519, 2015
|
|
4XBR
 
 | | In cellulo Crystal Structure of PAK4 in complex with Inka | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein FAM212A,Serine/threonine-protein kinase PAK 4 | | 著者 | Baskaran, Y, Ang, K.C, Anekal, P.V, Chan, W.L, Grimes, J.M, Manser, E, Robinson, R.C. | | 登録日 | 2014-12-17 | | 公開日 | 2015-12-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.94 Å) | | 主引用文献 | An in cellulo-derived structure of PAK4 in complex with its inhibitor Inka1
Nat Commun, 6, 2015
|
|
4XDS
 
 | | Deoxyguanosinetriphosphate Triphosphohydrolase from Escherichia coli with Nickel | | 分子名称: | Deoxyguanosinetriphosphate triphosphohydrolase, NICKEL (II) ION, SULFATE ION | | 著者 | Singh, D, Gawel, D, Itsko, M, Krahn, J.M, London, R.E, Schaaper, R.M. | | 登録日 | 2014-12-19 | | 公開日 | 2015-02-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.354 Å) | | 主引用文献 | Structure of Escherichia coli dGTP Triphosphohydrolase: A HEXAMERIC ENZYME WITH DNA EFFECTOR MOLECULES.
J.Biol.Chem., 290, 2015
|
|
2YJ7
 
 | | Crystal structure of a hyperstable protein from the Precambrian period | | 分子名称: | LPBCA THIOREDOXIN, SODIUM ION | | 著者 | Gavira, J.A, Ingles, A, Ibarra, B, Garcia-Ruiz, J.M. | | 登録日 | 2011-05-19 | | 公開日 | 2012-05-30 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Conservation of Protein Structure Over Four Billion Years
Structure, 21, 2013
|
|
2XWE
 
 | | X-RAY STRUCTURE OF ACID-BETA-GLUCOSIDASE WITH 5N,6S-(N'-(N-OCTYL)IMINO)-6-THIONOJIRIMYCIN IN THE ACTIVE SITE | | 分子名称: | (3Z,5S,6R,7S,8R,8aS)-3-(octylimino)hexahydro[1,3]thiazolo[3,4-a]pyridine-5,6,7,8-tetrol, GLUCOSYLCERAMIDASE, PHOSPHATE ION, ... | | 著者 | Brumshtein, B, Aguilar-Moncayo, M, Benito, J.M, Ortiz Mellet, C, Garcia Fernandez, J.M, Silman, I, Shaaltiel, Y, Sussman, J.L, Futerman, A.H. | | 登録日 | 2010-11-02 | | 公開日 | 2011-09-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Cyclodextrin-Mediated Crystallization of Acid Beta-Glucosidase in Complex with Amphiphilic Bicyclic Nojirimycin Analogues.
Org.Biomol.Chem., 9, 2011
|
|
2JVG
 
 | | Structure of C3-binding domain 4 of Staphylococcus aureus protein Sbi | | 分子名称: | IgG-binding protein SBI | | 著者 | Upadhyay, A, Burman, J, Clark, E.A, van den Elsen, J.M.H, Bagby, S. | | 登録日 | 2007-09-20 | | 公開日 | 2008-06-10 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure-function analysis of the C3 binding region of Staphylococcus aureus immune subversion protein Sbi.
J.Biol.Chem., 283, 2008
|
|
2HUO
 
 | | Crystal structure of mouse myo-inositol oxygenase in complex with substrate | | 分子名称: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, FE (III) ION, FORMIC ACID, ... | | 著者 | Brown, P.M, Caradoc-Davies, T.T, Dickson, J.M.J, Cooper, G.J.S, Loomes, K.M, Baker, E.N. | | 登録日 | 2006-07-27 | | 公開日 | 2006-09-26 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of a substrate complex of myo-inositol oxygenase, a di-iron oxygenase with a key role in inositol metabolism.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2JVH
 
 | | Structure of C3-binding domain 4 of S. aureus protein Sbi | | 分子名称: | IgG-binding protein SBI | | 著者 | Upadhyay, A, Burman, J, Clark, E.A, van den Elsen, J.M.H, Bagby, S. | | 登録日 | 2007-09-20 | | 公開日 | 2008-06-10 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure-function analysis of the C3 binding region of Staphylococcus aureus immune subversion protein Sbi.
J.Biol.Chem., 283, 2008
|
|
3C4O
 
 | | Crystal Structure of the SHV-1 Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) E73M/S130K/S146M complex | | 分子名称: | Beta-lactamase SHV-1, Beta-lactamase inhibitory protein, SULFATE ION | | 著者 | Reynolds, K.A, Hanes, M.S, Thomson, J.M, Antczak, A.J, Berger, J.M, Bonomo, R.A, Kirsch, J.F, Handel, T.M. | | 登録日 | 2008-01-30 | | 公開日 | 2008-05-27 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Computational redesign of the SHV-1 beta-lactamase/beta-lactamase inhibitor protein interface.
J.Mol.Biol., 382, 2008
|
|
3C9D
 
 | | Crystal structure of Vps75 | | 分子名称: | Vacuolar protein sorting-associated protein 75 | | 著者 | Berndsen, C.E, Tsubota, T, Lindner, S.E, Lee, S, Holton, J.M, Kaufman, P.D, Keck, J.L, Denu, J.M. | | 登録日 | 2008-02-15 | | 公開日 | 2008-08-12 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Molecular functions of the histone acetyltransferase chaperone complex Rtt109-Vps75
Nat.Struct.Mol.Biol., 15, 2008
|
|
2JOL
 
 | | Average NMR structure of the catalytic domain of guanine nucleotide exchange factor BopE from Burkholderia pseudomallei | | 分子名称: | Putative G-nucleotide exchange factor | | 著者 | Wu, H, Upadhyay, A, Williams, C, Galyov, E.E, van den Elsen, J.M.H, Bagby, S. | | 登録日 | 2007-03-14 | | 公開日 | 2007-03-27 | | 最終更新日 | 2023-12-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The guanine-nucleotide-exchange factor BopE from Burkholderia pseudomallei adopts a compact version of the Salmonella SopE/SopE2 fold and undergoes a closed-to-open conformational change upon interaction with Cdc42
Biochem.J., 411, 2008
|
|
2JOK
 
 | | NMR structure of the catalytic domain of guanine nucleotide exchange factor BopE from Burkholderia pseudomallei | | 分子名称: | Putative G-nucleotide exchange factor | | 著者 | Wu, H, Upadhyay, A, Williams, C, Galyov, E.E, van den Elsen, J.M.H, Bagby, S. | | 登録日 | 2007-03-14 | | 公開日 | 2007-09-18 | | 最終更新日 | 2023-12-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The guanine-nucleotide-exchange factor BopE from Burkholderia pseudomallei adopts a compact version of the Salmonella SopE/SopE2 fold and undergoes a closed-to-open conformational change upon interaction with Cdc42
Biochem.J., 411, 2008
|
|
1A94
 
 | | STRUCTURAL BASIS FOR SPECIFICITY OF RETROVIRAL PROTEASES | | 分子名称: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE | | 著者 | Wu, J, Adomat, J.M, Ridky, T.W, Louis, J.M, Leis, J, Harrison, R.W, Weber, I.T. | | 登録日 | 1998-04-16 | | 公開日 | 1999-01-13 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis for specificity of retroviral proteases.
Biochemistry, 37, 1998
|
|
1AOL
 
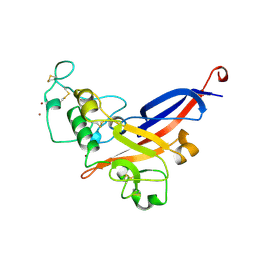 | | FRIEND MURINE LEUKEMIA VIRUS RECEPTOR-BINDING DOMAIN | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GP70, ZINC ION | | 著者 | Fass, D, Davey, R.A, Hamson, C.A, Kim, P.S, Cunningham, J.M, Berger, J.M. | | 登録日 | 1997-07-08 | | 公開日 | 1997-10-15 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 angstrom resolution.
Science, 277, 1997
|
|
1BAI
 
 | | Crystal structure of Rous sarcoma virus protease in complex with inhibitor | | 分子名称: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE | | 著者 | Wu, J, Adomat, J.M, Ridky, T.W, Louis, J.M, Leis, J, Harrison, R.W, Weber, I.T. | | 登録日 | 1998-04-17 | | 公開日 | 1999-01-13 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis for specificity of retroviral proteases.
Biochemistry, 37, 1998
|
|
1BYL
 
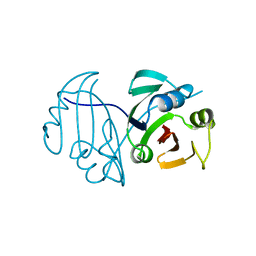 | |
2BF2
 
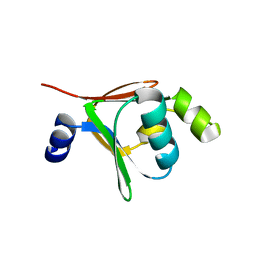 | | Crystal structure of native toluene-4-monooxygenase catalytic effector protein, T4moD | | 分子名称: | TOLUENE-4-MONOOXYGENASE SYSTEM PROTEIN D | | 著者 | Lountos, G.T, Mitchell, K.H, Studts, J.M, Fox, B.G, Orville, A.M. | | 登録日 | 2004-12-03 | | 公開日 | 2005-05-19 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structures and Functional Studies of T4Mod, the Toluene 4-Monooxygenase Catalytic Effector Protein
Biochemistry, 44, 2005
|
|
2BF3
 
 | | Crystal structure of a toluene 4-monooxygenase catalytic effector protein variant missing ten N-terminal residues (delta-N10 T4moD) | | 分子名称: | HEPTANE-1,2,3-TRIOL, TOLUENE-4-MONOOXYGENASE SYSTEM PROTEIN D | | 著者 | Lountos, G.T, Mitchell, K.H, Studts, J.M, Fox, B.G, Orville, A.M. | | 登録日 | 2004-12-03 | | 公開日 | 2005-05-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Crystal Structures and Functional Studies of T4Mod, the Toluene 4-Monooxygenase Catalytic Effector Protein
Biochemistry, 44, 2005
|
|
2BF5
 
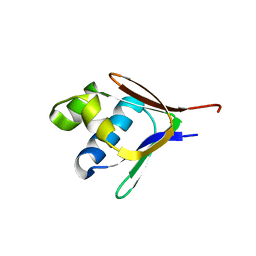 | | Crystal structure of a toluene 4-monooxygenase catalytic effector protein variant missing four N-terminal residues (delta-N4 T4moD) | | 分子名称: | TOLUENE-4-MONOOXYGENASE SYSTEM PROTEIN D | | 著者 | Lountos, G.T, Mitchell, K.H, Studts, J.M, Fox, B.G, Orville, A.M. | | 登録日 | 2004-12-03 | | 公開日 | 2005-05-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Crystal Structures and Functional Studies of T4Mod, the Toluene 4-Monooxygenase Catalytic Effector Protein
Biochemistry, 44, 2005
|
|
5ANY
 
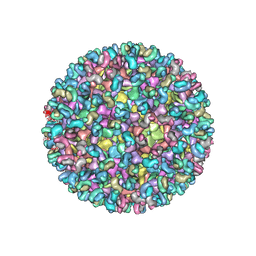 | | Electron cryo-microscopy of chikungunya virus in complex with neutralizing antibody Fab CHK265 | | 分子名称: | E1, E2, FAB, ... | | 著者 | Fox, J.M, Long, F, Edeling, M.A, Lin, H, Duijl-Richter, M, Fong, R.H, Kahle, K.M, Smit, J.M, Jin, J, Simmons, G, Doranz, B.J, Crowe, J.E, Fremont, D.H, Rossmann, M.G, Diamond, M.S. | | 登録日 | 2015-09-08 | | 公開日 | 2015-11-25 | | 最終更新日 | 2018-10-03 | | 実験手法 | ELECTRON MICROSCOPY (16.9 Å) | | 主引用文献 | Broadly Neutralizing Alphavirus Antibodies Bind an Epitope on E2 and Inhibit Entry and Egress.
Cell(Cambridge,Mass.), 163, 2015
|
|
1ATP
 
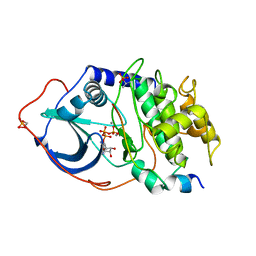 | | 2.2 angstrom refined crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MNATP and a peptide inhibitor | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, PEPTIDE INHIBITOR PKI(5-24), ... | | 著者 | Zheng, J, Trafny, E.A, Knighton, D.R, Xuong, N.-H, Taylor, S.S, Teneyck, L.F, Sowadski, J.M. | | 登録日 | 1993-01-08 | | 公開日 | 1993-04-15 | | 最終更新日 | 2019-08-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | 2.2 A refined crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MnATP and a peptide inhibitor.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1HXS
 
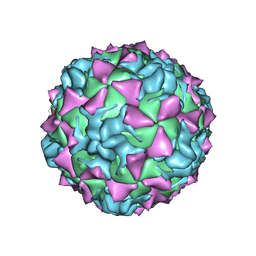 | | CRYSTAL STRUCTURE OF MAHONEY STRAIN OF POLIOVIRUS AT 2.2A RESOLUTION | | 分子名称: | GENOME POLYPROTEIN, COAT PROTEIN VP1, COAT PROTEIN VP2, ... | | 著者 | Miller, S.T, Hogle, J.M, Filman, D.J. | | 登録日 | 2001-01-16 | | 公開日 | 2002-01-16 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Ab initio phasing of high-symmetry macromolecular complexes: successful phasing of authentic poliovirus data to 3.0 A resolution.
J.Mol.Biol., 307, 2001
|
|
6PTL
 
 | |
3OED
 
 | |
