1FQE
 
 | | CRYSTAL STRUCTURES OF MUTANT (K206A) THAT ABOLISH THE DILYSINE INTERACTION IN THE N-LOBE OF HUMAN TRANSFERRIN | | 分子名称: | CARBONATE ION, FE (III) ION, POTASSIUM ION, ... | | 著者 | Nurizzo, D, Baker, H.M, Baker, E.N. | | 登録日 | 2000-09-04 | | 公開日 | 2001-05-16 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structures and iron release properties of mutants (K206A and K296A) that abolish the dilysine interaction in the N-lobe of human transferrin.
Biochemistry, 40, 2001
|
|
8F7F
 
 | | The condensation domain of surfactin A synthetase C in space group P43212 | | 分子名称: | GLYCEROL, Surfactin synthetase | | 著者 | Frota, N, Pistofidis, A, Folger, I.B, Hilvert, D, Schmeing, T.M. | | 登録日 | 2022-11-18 | | 公開日 | 2023-11-22 | | 最終更新日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | High-throughput reprogramming of an NRPS condensation domain.
Nat.Chem.Biol., 20, 2024
|
|
6P7W
 
 | | Structure of the K. lactis CBF3 core - Ndc10 D1 complex | | 分子名称: | Cep3, Ctf13, Ndc10, ... | | 著者 | Lee, P.D, Wei, H, Tan, D, Harrison, S.C. | | 登録日 | 2019-06-06 | | 公開日 | 2019-09-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structure of the Centromere Binding Factor 3 Complex from Kluyveromyces lactis.
J.Mol.Biol., 431, 2019
|
|
1FF5
 
 | | STRUCTURE OF E-CADHERIN DOUBLE DOMAIN | | 分子名称: | CALCIUM ION, EPITHELIAL CADHERIN | | 著者 | Pertz, O, Bozic, D, Koch, A.W, Fauser, C, Brancaccio, A, Engel, J. | | 登録日 | 2000-07-25 | | 公開日 | 2000-08-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.93 Å) | | 主引用文献 | A new crystal structure, Ca2+ dependence and mutational analysis reveal molecular details of E-cadherin homoassociation.
EMBO J., 18, 1999
|
|
8C6C
 
 | | Light SFX structure of D.m(6-4)photolyase at 300ps time delay | | 分子名称: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | 著者 | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | 登録日 | 2023-01-11 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
4XS1
 
 | | Salmonella typhimurium AhpC T43V mutant | | 分子名称: | Alkyl hydroperoxide reductase subunit C, CHLORIDE ION, POTASSIUM ION, ... | | 著者 | Perkins, A, Nelson, K, Parsonage, D, Poole, L, Karplus, P.A. | | 登録日 | 2015-01-21 | | 公開日 | 2016-01-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Experimentally Dissecting the Origins of Peroxiredoxin Catalysis.
Antioxid.Redox Signal., 28, 2018
|
|
8C6H
 
 | | Light SFX structure of D.m(6-4)photolyase at 2ps time delay | | 分子名称: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | 著者 | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | 登録日 | 2023-01-11 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
4XZ4
 
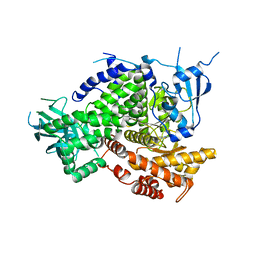 | | Structure of PI3K gamma in complex with an inhibitor | | 分子名称: | N-[5-(6-methoxypyrazin-2-yl)-4,5,6,7-tetrahydro[1,3]thiazolo[5,4-c]pyridin-2-yl]acetamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | 著者 | Collier, P.N, Messersmith, D, Le Tiran, A, Bandarage, U.K, Boucher, C, Come, J, Cottrell, K.M, Damagnez, V, Doran, J.D, Griffith, J.P, Khare-Pandit, S, Krueger, E.B, Ledeboer, M.W, Ledford, B, Liao, Y, Mahajan, S, Moody, C.S, Wang, T, Xu, J, Aronov, A.M. | | 登録日 | 2015-02-03 | | 公開日 | 2016-02-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure of PI3K gamma in complex with an inhibitor
To Be Published
|
|
8C1U
 
 | | SFX structure of D.m(6-4)photolyase | | 分子名称: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | 著者 | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | 登録日 | 2022-12-21 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C69
 
 | | Light SFX structure of D.m(6-4)photolyase at 100 microsecond time delay | | 分子名称: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | 著者 | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | 登録日 | 2023-01-11 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C6A
 
 | | Light SFX structure of D.m(6-4)photolyase at 1ps time delay | | 分子名称: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | 著者 | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | 登録日 | 2023-01-11 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
4MB9
 
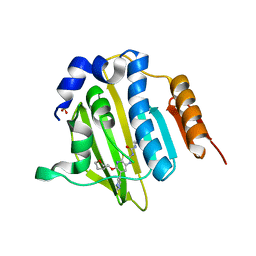 | | Structure of Streptococcus pneumonia ParE in complex with AZ13102335 | | 分子名称: | 1-ethyl-3-{6-(pyrimidin-5-yl)-5-[(3R)-tetrahydrofuran-3-ylmethoxy][1,3]thiazolo[5,4-b]pyridin-2-yl}urea, DNA topoisomerase IV, B subunit, ... | | 著者 | Ogg, D, Tucker, J. | | 登録日 | 2013-08-19 | | 公開日 | 2013-10-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Thiazolopyridine Ureas as Novel Antitubercular Agents Acting through Inhibition of DNA Gyrase B.
J.Med.Chem., 56, 2013
|
|
4MBC
 
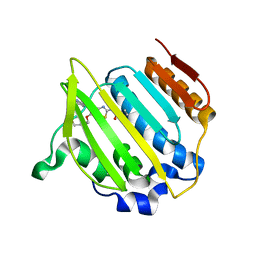 | | Structure of Streptococcus pneumonia ParE in complex with AZ13053807 | | 分子名称: | 1-{5-[2-(morpholin-4-yl)ethoxy]-6-(pyridin-3-yl)[1,3]thiazolo[5,4-b]pyridin-2-yl}-3-prop-2-en-1-ylurea, DNA topoisomerase IV, B subunit | | 著者 | Ogg, D, Tucker, J. | | 登録日 | 2013-08-19 | | 公開日 | 2013-10-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Thiazolopyridine Ureas as Novel Antitubercular Agents Acting through Inhibition of DNA Gyrase B.
J.Med.Chem., 56, 2013
|
|
8C6B
 
 | | Light SFX structure of D.m(6-4)photolyase at 20ps time delay | | 分子名称: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | 著者 | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | 登録日 | 2023-01-11 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C6F
 
 | | Light SFX structure of D.m(6-4)photolyase at 400fs time delay | | 分子名称: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | 著者 | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | 登録日 | 2023-01-11 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8ENX
 
 | |
8ENZ
 
 | |
6P0S
 
 | | Crystal structure of ternary DNA complex "FX2" containing E. coli Fis and phage lambda Xis | | 分子名称: | DNA (27-MER), FX1-2, DNA-binding protein Fis, ... | | 著者 | Hancock, S.P, Cascio, D, Johnson, R.C. | | 登録日 | 2019-05-17 | | 公開日 | 2019-06-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Cooperative DNA binding by proteins through DNA shape complementarity.
Nucleic Acids Res., 47, 2019
|
|
8ENY
 
 | |
8ENS
 
 | |
8CIL
 
 | |
8EO0
 
 | |
8ENW
 
 | |
6P4A
 
 | |
8C7C
 
 | | Double mutant V(M84)C/A(L278)C structure of Photosynthetic Reaction Center From Cereibacter sphaeroides strain RV | | 分子名称: | 1,2-ETHANEDIOL, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | 著者 | Gabdulkhakov, A, Selikhanov, G, Fufina, T, Vasilieva, L, Atamas, A, Uhimchuk, D. | | 登録日 | 2023-01-14 | | 公開日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Stabilization of Cereibacter sphaeroides Photosynthetic Reaction Center by the Introduction of Disulfide Bonds.
Membranes (Basel), 13, 2023
|
|
