8EV2
 
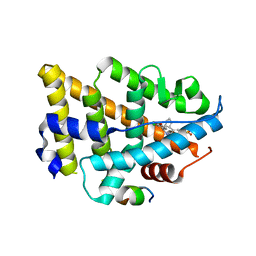 | | Dual Modulators | | 分子名称: | (3aS,4R,9bR)-4-(2-chloro-4-hydroxyphenyl)-2,3,3a,4,5,9b-hexahydro-1H-cyclopenta[c]quinoline-8-sulfonamide, (3~{a}~{R},4~{S},9~{b}~{S})-4-(2-chloranyl-4-oxidanyl-phenyl)-2,3,3~{a},4,5,9~{b}-hexahydro-1~{H}-cyclopenta[c]quinoline-8-sulfonamide, Estrogen receptor, ... | | 著者 | Tinivella, A, Nwachukwu, J.C, Angeli, A, Foschi, F, Benatti, A.L, Pinzi, L, Izard, T, Ferraroni, M, Rangarajan, E.S, Christodoulou, M, Passarella, D, Supuran, C, Nettles, K.W, Rastelli, G. | | 登録日 | 2022-10-19 | | 公開日 | 2022-12-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Design, synthesis, biological evaluation and crystal structure determination of dual modulators of carbonic anhydrases and estrogen receptors.
Eur.J.Med.Chem., 246, 2022
|
|
6QM9
 
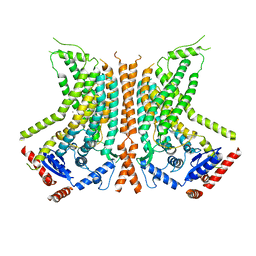 | | Cryo-EM structure of calcium-bound nhTMEM16 lipid scramblase in nanodisc (open state) | | 分子名称: | CALCIUM ION, Predicted protein | | 著者 | Kalienkova, V, Clerico Mosina, V, Bryner, L, Oostergetel, G.T, Dutzler, R, Paulino, C. | | 登録日 | 2019-02-01 | | 公開日 | 2019-03-06 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Elife, 8, 2019
|
|
3ZLK
 
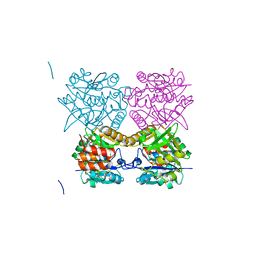 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | 分子名称: | CHLORIDE ION, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, N-(6-AMINO-1-BENZYL-2,4-DIOXO-1,2,3,4-TETRAHYDROPYRIMIDIN-5-YL)BENZENESULFONAMIDE | | 著者 | Alphey, M.S, Pirrie, L, Torrie, L.S, Gardiner, M, Sarkar, A, Brenk, R, Westwood, N.J, Gray, D, Naismith, J.H. | | 登録日 | 2013-02-01 | | 公開日 | 2013-02-20 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.952 Å) | | 主引用文献 | Allosteric competitive inhibitors of the glucose-1-phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa.
ACS Chem. Biol., 8, 2013
|
|
8F41
 
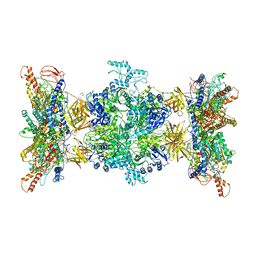 | | 3-methylcrotonyl-CoA carboxylase in filament, alpha-subunit centered | | 分子名称: | 3-methylcrotonyl-CoA carboxylase, alpha-subunit, beta-subunit, ... | | 著者 | Hu, J.J, Lee, J.K.J, Liu, Y.T, Yu, C, Huang, L, Afasizheva, I, Afasizhev, R, Zhou, Z.H. | | 登録日 | 2022-11-10 | | 公開日 | 2023-01-11 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Discovery, structure, and function of filamentous 3-methylcrotonyl-CoA carboxylase.
Structure, 31, 2023
|
|
4B23
 
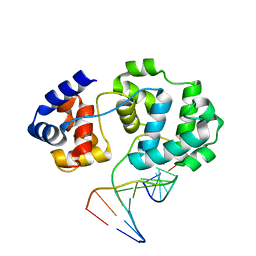 | | Unprecedented sculpting of DNA at abasic sites by DNA glycosylase homolog Mag2 | | 分子名称: | 5'-D(*CP*GP*AP*TP*TP*GP*GP*TP*AP*GP*CP)-3', 5'-D(*GP*CP*TP*AP*CP*(3DR)P*AP*AP*TP*CP*GP)-3', MAG2, ... | | 著者 | Dalhus, B, Nilsen, L, Korvald, H, Huffman, J, Forstrom, R.J, McMurray, C.T, Alseth, I, Tainer, J.A, Bjoras, M. | | 登録日 | 2012-07-12 | | 公開日 | 2013-01-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Sculpting of DNA at Abasic Sites by DNA Glycosylase Homolog Mag2.
Structure, 21, 2013
|
|
6QLY
 
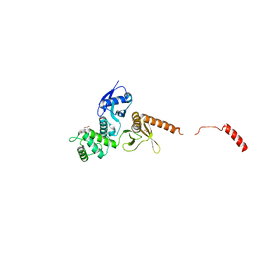 | | IDOL FERM domain | | 分子名称: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase MYLIP, SULFATE ION | | 著者 | Martinelli, L, Sixma, T.K. | | 登録日 | 2019-02-01 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural analysis of the LDL receptor-interacting FERM domain in the E3 ubiquitin ligase IDOL reveals an obscured substrate-binding site.
J.Biol.Chem., 295, 2020
|
|
6QM4
 
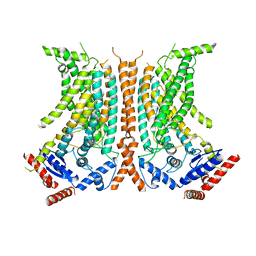 | | Cryo-EM structure of calcium-free nhTMEM16 lipid scramblase in nanodisc | | 分子名称: | Predicted protein | | 著者 | Kalienkova, V, Clerico Mosina, V, Bryner, L, Oostergetel, G.T, Dutzler, R, Paulino, C. | | 登録日 | 2019-02-01 | | 公開日 | 2019-03-06 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Elife, 8, 2019
|
|
4B85
 
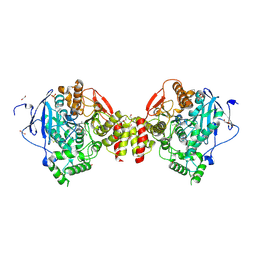 | | Mus musculus Acetylcholinesterase in complex with 4-Chloranyl-N-(2- diethylamino-ethyl)-benzenesulfonamide | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-chloranyl-N-[2-(diethylamino)ethyl]benzenesulfonamide, ACETYLCHOLINESTERASE, ... | | 著者 | Andersson, C.D, Forsgren, N, Akfur, C, Allgardsson, A, Berg, L, Qian, W, Ekstrom, F, Linusson, A. | | 登録日 | 2012-08-24 | | 公開日 | 2013-09-04 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Divergent Structure-Activity Relationships of Structurally Similar Acetylcholinesterase Inhibitors.
J.Med.Chem., 56, 2013
|
|
4BB9
 
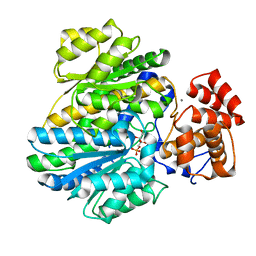 | | Crystal structure of glucokinase regulatory protein complexed to fructose-1-phosphate | | 分子名称: | 1-O-phosphono-beta-D-fructopyranose, CALCIUM ION, GLUCOKINASE REGULATORY PROTEIN | | 著者 | Pautsch, A, Stadler, N, Loehle, A, Lenter, M, Rist, W, Berg, A, Glocker, L, Nar, H, Reinert, D, Heckel, A, Schnapp, G, Kauschke, S.G. | | 登録日 | 2012-09-21 | | 公開日 | 2013-05-15 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Crystal Structure of Glucokinase Regulatory Protein.
Biochemistry, 52, 2013
|
|
8E6D
 
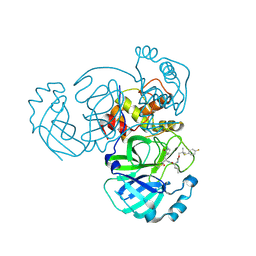 | | Crystal structure of MERS 3CL protease in complex with a p-fluorophenyl dimethyl sulfane inhibitor | | 分子名称: | (1R,2S)-2-{[N-({2-[(4-fluorophenyl)sulfanyl]-2-methylpropoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, Orf1a protein | | 著者 | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | 登録日 | 2022-08-22 | | 公開日 | 2022-09-07 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
8E61
 
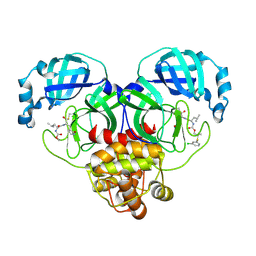 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a m-chlorophenyl dimethyl sulfane inhibitor | | 分子名称: | (1R,2S)-2-{[N-({2-[(3-chlorophenyl)sulfanyl]-2-methylpropoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({2-[(3-chlorophenyl)sulfanyl]-2-methylpropoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | 著者 | Lovell, S, Liu, L, Battaile, K.P, Madden, T.K, Groutas, W.C. | | 登録日 | 2022-08-22 | | 公開日 | 2022-09-07 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
8E6B
 
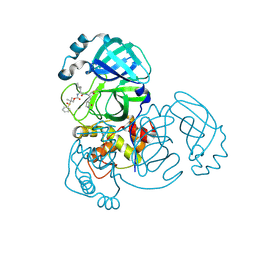 | | Crystal structure of MERS 3CL protease in complex with a dimethyl sulfinyl benzene inhibitor | | 分子名称: | (2~{S})-2-[[(2~{S})-4-methyl-2-[[2-methyl-2-[oxidanyl(phenyl)-$l^{3}-sulfanyl]propoxy]carbonylamino]pentanoyl]amino]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propane-1-sulfonic acid, N~2~-(ethoxycarbonyl)-N-{(1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-1-sulfanylpropan-2-yl}-L-leucinamide, Orf1a protein | | 著者 | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | 登録日 | 2022-08-22 | | 公開日 | 2022-09-07 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
4AWN
 
 | | Structure of recombinant human DNase I (rhDNaseI) in complex with Magnesium and Phosphate. | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Parsiegla, G, Noguere, C, Santell, L, Lazarus, R.A, Bourne, Y. | | 登録日 | 2012-06-04 | | 公開日 | 2013-01-09 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | The Structure of Human DNase I Bound to Magnesium and Phosphate Ions Points to a Catalytic Mechanism Common to Members of the DNase I-Like Superfamily.
Biochemistry, 51, 2012
|
|
6QZ8
 
 | | Structure of Mcl-1 in complex with compound 10d | | 分子名称: | (2~{R})-2-[5-(3-chloranyl-2-methyl-4-oxidanyl-phenyl)-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]oxy-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | 著者 | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | 登録日 | 2019-03-11 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
8E6E
 
 | | Crystal structure of MERS 3CL protease in complex with a phenyl sulfane inhibitor | | 分子名称: | (1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-[(N-{[2-(phenylsulfanyl)ethoxy]carbonyl}-L-leucyl)amino]propane-1-sulfonic acid, 2-phenylsulfanylethyl ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, Orf1a protein | | 著者 | Liu, L, Lovell, S, Battaile, K.P, Madden, T.K, Groutas, W.C. | | 登録日 | 2022-08-22 | | 公開日 | 2022-09-07 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
4AVF
 
 | | Crystal structure of Pseudomonas aeruginosa inosine 5'-monophosphate dehydrogenase | | 分子名称: | INOSINE-5'-MONOPHOSPHATE DEHYDROGENASE | | 著者 | McMahon, S.A, Moynie, L, Liu, H, Duthie, F, Naismith, J.H. | | 登録日 | 2012-05-25 | | 公開日 | 2013-01-09 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery
Acta Crystallogr.,Sect.F, 69, 2013
|
|
8E6C
 
 | | Crystal structure of MERS 3CL protease in complex with a m-fluorophenyl dimethyl sulfane inhibitor | | 分子名称: | Orf1a protein, [2-(3-fluorophenyl)sulfanyl-2-methyl-propyl] ~{N}-[(2~{S})-1-[[3-[(3~{S})-2-$l^{3}-oxidanylidenepyrrolidin-3-yl]-1-$l^{1}-oxidanylsulfonyl-1-oxidanyl-propan-2-yl]-$l^{2}-azanyl]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | | 著者 | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | 登録日 | 2022-08-22 | | 公開日 | 2022-09-07 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
8E69
 
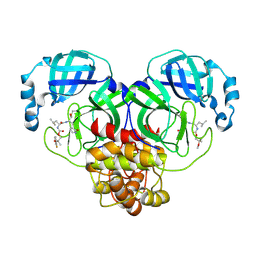 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a m-fluorodimethyl oxybenzene inhibitor | | 分子名称: | (1R,2S)-2-[(N-{[2-(3-fluorophenoxy)-2-methylpropoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | 著者 | Lovell, S, Liu, L, Battaile, K.P, Miller, M.J, Groutas, W.C. | | 登録日 | 2022-08-22 | | 公開日 | 2022-09-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
6ARY
 
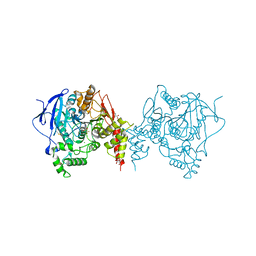 | | Crystal structure of an insecticide-resistant acetylcholinesterase mutant from the malaria vector Anopheles gambiae in complex with a difluoromethyl ketone inhibitor | | 分子名称: | (1S)-2,2-difluoro-1-[1-(pentan-3-yl)-1H-pyrazol-4-yl]ethan-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | 著者 | Cheung, J, Mahmood, A, Kalathur, R, Lixuan, L, Carlier, P.R. | | 登録日 | 2017-08-23 | | 公開日 | 2018-01-10 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.257 Å) | | 主引用文献 | Structure of the G119S Mutant Acetylcholinesterase of the Malaria Vector Anopheles gambiae Reveals Basis of Insecticide Resistance.
Structure, 26, 2018
|
|
6QV5
 
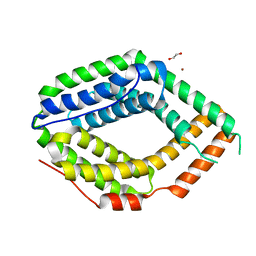 | |
4B80
 
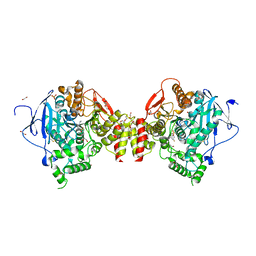 | | Mus musculus Acetylcholinesterase in complex with N-(2-Diethylamino-ethyl)-1-(4-fluoro-phenyl)-methanesulfonamide | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | 著者 | Andersson, C.D, Forsgren, N, Akfur, C, Allgardsson, A, Berg, L, Qian, W, Ekstrom, F, Linusson, A. | | 登録日 | 2012-08-24 | | 公開日 | 2013-09-04 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Divergent structure-activity relationships of structurally similar acetylcholinesterase inhibitors.
J. Med. Chem., 56, 2013
|
|
4AS2
 
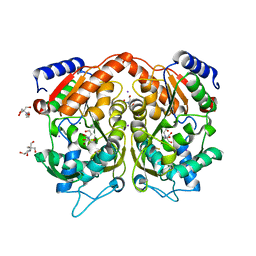 | | Pseudomonas Aeruginosa Phosphorylcholine Phosphatase. Monoclinic form | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, IODIDE ION, ... | | 著者 | Infantes, L, Otero, L.H, Albert, A. | | 登録日 | 2012-04-27 | | 公開日 | 2012-08-22 | | 最終更新日 | 2019-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | The Structural Domains of Pseudomonas Aeruginosa Phosphorylcholine Phosphatase Cooperate in Substrate Hydrolysis: 3D Structure and Enzymatic Mechanism.
J.Mol.Biol., 423, 2012
|
|
6QYP
 
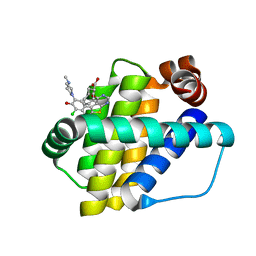 | | Structure of Mcl-1 in complex with compound 13 | | 分子名称: | (2~{R})-2-[5-[3-chloranyl-2-methyl-5-(4-methylpiperazin-1-yl)-4-oxidanyl-phenyl]-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]oxy-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | 著者 | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | 登録日 | 2019-03-09 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QZ5
 
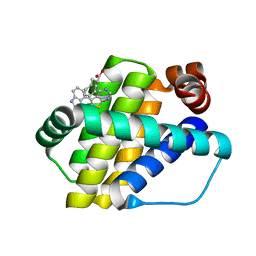 | | Structure of Mcl-1 in complex with compound 8a | | 分子名称: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | 著者 | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | 登録日 | 2019-03-11 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
4B21
 
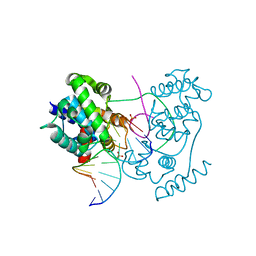 | | Unprecedented sculpting of DNA at abasic sites by DNA glycosylase homolog Mag2 | | 分子名称: | 5'-D(*CP*GP*AP*TP*CP*GP*GP*TP*AP*GP)-3', 5'-D(*GP*CP*TP*AP*CP*3DRP*GP*AP*TP*CP*GP)-3', PHOSPHATE ION, ... | | 著者 | Dalhus, B, Nilsen, L, Korvald, H, Huffman, J, Forstrom, R.J, McMurray, C.T, Alseth, I, Tainer, J.A, Bjoras, M. | | 登録日 | 2012-07-12 | | 公開日 | 2013-01-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Sculpting of DNA at Abasic Sites by DNA Glycosylase Homolog Mag2.
Structure, 21, 2013
|
|
