5X7Q
 
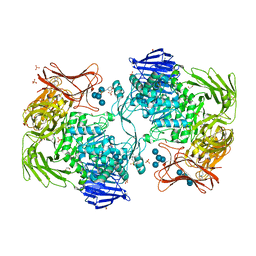 | | Crystal structure of Paenibacillus sp. 598K alpha-1,6-glucosyltransferase complexed with maltohexaose | | 分子名称: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | 著者 | Fujimoto, Z, Kishine, N, Suzuki, N, Momma, M, Ichinose, H, Kimura, A, Funane, K. | | 登録日 | 2017-02-27 | | 公開日 | 2017-07-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Carbohydrate-binding architecture of the multi-modular alpha-1,6-glucosyltransferase from Paenibacillus sp. 598K, which produces alpha-1,6-glucosyl-alpha-glucosaccharides from starch
Biochem. J., 474, 2017
|
|
2D2P
 
 | | The solution structure of micelle-bound peptide | | 分子名称: | Pituitary adenylate cyclase activating polypeptide-38 | | 著者 | Tateishi, Y, Jee, J.G, Inooka, H, Tochio, H, Hiroaki, H, Shirakawa, M. | | 登録日 | 2005-09-14 | | 公開日 | 2006-09-26 | | 最終更新日 | 2022-03-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of micelle-bound peptide
To be Published
|
|
3WR7
 
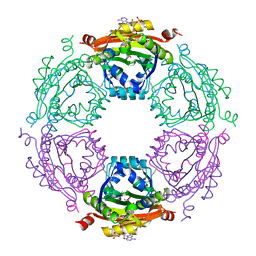 | | Crystal Structure of Spermidine Acetyltransferase from Escherichia coli | | 分子名称: | COENZYME A, SPERMIDINE, Spermidine N1-acetyltransferase | | 著者 | Sugiyama, S, Ishikawa, S, Tomitori, S, Niiyama, M, Hirose, M, Miyazaki, Y, Higashi, K, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Kashiwagi, K, Igarashi, K, Matsumura, H. | | 登録日 | 2014-02-20 | | 公開日 | 2015-09-02 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Molecular mechanism underlying promiscuous polyamine recognition by spermidine acetyltransferase
Int.J.Biochem.Cell Biol., 76, 2016
|
|
2EXD
 
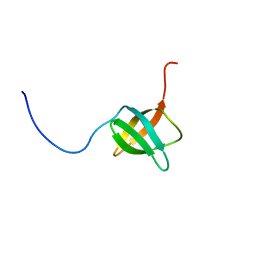 | | The solution structure of the C-terminal domain of a nfeD homolog from Pyrococcus horikoshii | | 分子名称: | nfeD short homolog | | 著者 | Kuwahara, Y, Ohno, A, Morii, T, Tochio, H, Shirakawa, M, Hiroaki, H. | | 登録日 | 2005-11-08 | | 公開日 | 2006-12-12 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of the C-terminal domain of NfeD reveals a novel membrane-anchored OB-fold.
Protein Sci., 17, 2008
|
|
6I37
 
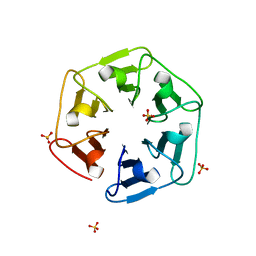 | | Crystal structure of nv1Pizza6-AYW, a circularly permuted designer protein | | 分子名称: | SULFATE ION, nv1Pizza6-AYW | | 著者 | Mylemans, B, Noguchi, H, Deridder, E, Voet, A.R.D. | | 登録日 | 2018-11-05 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Influence of circular permutations on the structure and stability of a six-fold circular symmetric designer protein.
Protein Sci., 2020
|
|
6I39
 
 | | Crystal structure of v31Pizza6-AYW, a circularly permuted designer protein | | 分子名称: | MAGNESIUM ION, v31Pizza6-AYW | | 著者 | Mylemans, B, Noguchi, H, Deridder, E, Voet, A.R.D. | | 登録日 | 2018-11-05 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Influence of circular permutations on the structure and stability of a six-fold circular symmetric designer protein.
Protein Sci., 2020
|
|
3VUS
 
 | | Escherichia coli PgaB N-terminal domain | | 分子名称: | ACETATE ION, MERCURY (II) ION, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase, ... | | 著者 | Nishiyama, T, Noguchi, H, Yoshida, H, Park, S.-Y, Tame, J.R.H. | | 登録日 | 2012-07-05 | | 公開日 | 2012-11-14 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | The structure of the deacetylase domain of Escherichia coli PgaB, an enzyme required for biofilm formation: a circularly permuted member of the carbohydrate esterase 4 family
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3W6H
 
 | | Crystal structure of 19F probe-labeled hCAI in complex with acetazolamide | | 分子名称: | 1-(2-ethoxyethoxy)-3,5-bis(trifluoromethyl)benzene, 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase 1, ... | | 著者 | Takaoka, Y, Kioi, Y, Morito, A, Otani, J, Arita, K, Ashihara, E, Ariyoshi, M, Tochio, H, Shirakawa, M, Hamachi, I. | | 登録日 | 2013-02-14 | | 公開日 | 2013-03-13 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.964 Å) | | 主引用文献 | Quantitative Comparison of Protein Dynamics in Live Cells and In Vitro by In-Cell 19F-NMR
To be published
|
|
3WN2
 
 | | Crystal Structure of Streptomyces coelicolor alpha-L-arabinofuranosidase in complex with xylohexaose | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Fujimoto, Z, Maehara, T, Ichinose, H, Michikawa, M, Harazono, K, Kaneko, S. | | 登録日 | 2013-11-29 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure and characterization of the glycoside hydrolase family 62 alpha-L-arabinofuranosidase from Streptomyces coelicolor
J.Biol.Chem., 289, 2014
|
|
3W6I
 
 | | Crystal structure of 19F probe-labeled hCAI | | 分子名称: | 1-(2-ethoxyethoxy)-3,5-bis(trifluoromethyl)benzene, Carbonic anhydrase 1, ZINC ION | | 著者 | Takaoka, Y, Kioi, Y, Morito, A, Otani, J, Arita, K, Ashihara, E, Ariyoshi, M, Tochio, H, Shirakawa, M, Hamachi, I. | | 登録日 | 2013-02-14 | | 公開日 | 2013-03-13 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.693 Å) | | 主引用文献 | Quantitative Comparison of Protein Dynamics in Live Cells and In Vitro by In-Cell 19F-NMR
To be published
|
|
3WMY
 
 | | Crystal Structure of Streptomyces coelicolor alpha-L-arabinofuranosidase | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Fujimoto, Z, Maehara, T, Ichinose, H, Michikawa, M, Harazono, K, Kaneko, S. | | 登録日 | 2013-11-29 | | 公開日 | 2014-02-05 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal structure and characterization of the glycoside hydrolase family 62 alpha-L-arabinofuranosidase from Streptomyces coelicolor
J.Biol.Chem., 289, 2014
|
|
5YPP
 
 | | Crystal structure of IlvN.Val-1a | | 分子名称: | ACETATE ION, Acetolactate synthase isozyme 1 small subunit, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Sarma, S.P, Bansal, A, Schindelin, H, Demeler, B. | | 登録日 | 2017-11-02 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystallographic Structures of IlvN·Val/Ile Complexes: Conformational Selectivity for Feedback Inhibition of Aceto Hydroxy Acid Synthases.
Biochemistry, 58, 2019
|
|
5YPW
 
 | | Crystal structure of IlvN.Val-1b | | 分子名称: | Acetolactate synthase isozyme 1 small subunit, VALINE | | 著者 | Sarma, S.P, Bansal, A, Schindelin, H, Demeler, B. | | 登録日 | 2017-11-03 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystallographic Structures of IlvN·Val/Ile Complexes: Conformational Selectivity for Feedback Inhibition of Aceto Hydroxy Acid Synthases.
Biochemistry, 58, 2019
|
|
3WMZ
 
 | | Crystal Structure of Streptomyces coelicolor alpha-L-arabinofuranosidase ethylmercury derivative | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ETHYL MERCURY ION, ... | | 著者 | Fujimoto, Z, Maehara, T, Ichinose, H, Michikawa, M, Harazono, K, Kaneko, S. | | 登録日 | 2013-11-29 | | 公開日 | 2014-02-05 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure and characterization of the glycoside hydrolase family 62 alpha-L-arabinofuranosidase from Streptomyces coelicolor
J.Biol.Chem., 289, 2014
|
|
3WN1
 
 | | Crystal Structure of Streptomyces coelicolor alpha-L-arabinofuranosidase in complex with xylotriose | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Fujimoto, Z, Maehara, T, Ichinose, H, Michikawa, M, Harazono, K, Kaneko, S. | | 登録日 | 2013-11-29 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure and characterization of the glycoside hydrolase family 62 alpha-L-arabinofuranosidase from Streptomyces coelicolor
J.Biol.Chem., 289, 2014
|
|
3WO2
 
 | | Crystal structure of human interleukin-18 | | 分子名称: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Interleukin-18, SULFATE ION | | 著者 | Tsutsumi, N, Kimura, T, Arita, K, Ariyoshi, M, Ohnishi, H, Kondo, N, Shirakawa, M, Kato, Z, Tochio, H. | | 登録日 | 2013-12-19 | | 公開日 | 2014-12-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | The structural basis for receptor recognition of human interleukin-18
Nat Commun, 5, 2014
|
|
3WO4
 
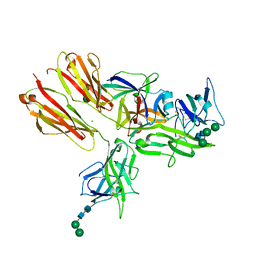 | | Crystal structure of the IL-18 signaling ternary complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Tsutsumi, N, Kimura, T, Arita, K, Ariyoshi, M, Ohnishi, H, Kondo, N, Shirakawa, M, Kato, Z, Tochio, H. | | 登録日 | 2013-12-19 | | 公開日 | 2014-12-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | The structural basis for receptor recognition of human interleukin-18
Nat Commun, 5, 2014
|
|
5YPY
 
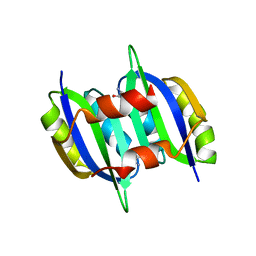 | | Crystal structure of IlvN. Val-1c | | 分子名称: | Acetolactate synthase isozyme 1 small subunit, VALINE | | 著者 | Sarma, S.P, Bansal, A, Schindelin, H, Demeler, B. | | 登録日 | 2017-11-04 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.966 Å) | | 主引用文献 | Crystallographic Structures of IlvN·Val/Ile Complexes: Conformational Selectivity for Feedback Inhibition of Aceto Hydroxy Acid Synthases.
Biochemistry, 58, 2019
|
|
3WKJ
 
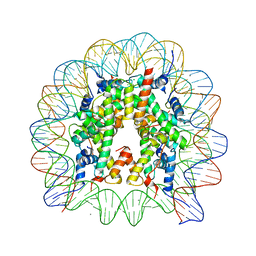 | | The nucleosome containing human TSH2B | | 分子名称: | CHLORIDE ION, DNA (145-MER), Histone H2A type 1-B/E, ... | | 著者 | Urahama, T, Horikoshi, N, Osakabe, A, Tachiwana, H, Kurumizaka, H. | | 登録日 | 2013-10-22 | | 公開日 | 2014-04-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of human nucleosome containing the testis-specific histone variant TSH2B.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
3WN0
 
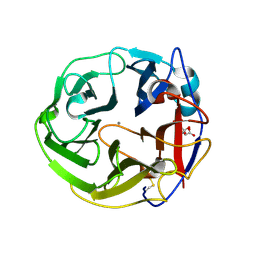 | | Crystal Structure of Streptomyces coelicolor alpha-L-arabinofuranosidase in complex with L-arabinose | | 分子名称: | CALCIUM ION, CHLORIDE ION, CITRIC ACID, ... | | 著者 | Fujimoto, Z, Maehara, T, Ichinose, H, Michikawa, M, Harazono, K, Kaneko, S. | | 登録日 | 2013-11-29 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure and characterization of the glycoside hydrolase family 62 alpha-L-arabinofuranosidase from Streptomyces coelicolor
J.Biol.Chem., 289, 2014
|
|
3WO3
 
 | | Crystal structure of IL-18 in complex with IL-18 receptor alpha | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Tsutsumi, N, Kimura, T, Arita, K, Ariyoshi, M, Ohnishi, H, Kondo, N, Shirakawa, M, Kato, Z, Tochio, H. | | 登録日 | 2013-12-19 | | 公開日 | 2014-12-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | The structural basis for receptor recognition of human interleukin-18
Nat Commun, 5, 2014
|
|
5Z20
 
 | | The ternary structure of D-lactate dehydrogenase from Pseudomonas aeruginosa with NADH and oxamate | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, D-lactate dehydrogenase (Fermentative), DI(HYDROXYETHYL)ETHER, ... | | 著者 | Furukawa, N, Miyanaga, A, Nakajima, M, Taguchi, H. | | 登録日 | 2017-12-28 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural Basis of Sequential Allosteric Transitions in Tetrameric d-Lactate Dehydrogenases from Three Gram-Negative Bacteria.
Biochemistry, 57, 2018
|
|
6HSO
 
 | | Crystal structure of the ternary complex of GephE-ADP-Glycine receptor derived peptide | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,1'-benzene-1,4-diylbis(1H-pyrrole-2,5-dione), ACETATE ION, ... | | 著者 | Kasaragod, V.B, Schindelin, H. | | 登録日 | 2018-10-01 | | 公開日 | 2019-01-16 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Elucidating the Molecular Basis for Inhibitory Neurotransmission Regulation by Artemisinins.
Neuron, 101, 2019
|
|
5Z1Z
 
 | | The apo-structure of D-lactate dehydrogenase from Escherichia coli | | 分子名称: | D-isomer specific 2-hydroxyacid dehydrogenase NAD-binding, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION | | 著者 | Furukawa, N, Miyanaga, A, Nakajima, M, Taguchi, H. | | 登録日 | 2017-12-28 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Structural Basis of Sequential Allosteric Transitions in Tetrameric d-Lactate Dehydrogenases from Three Gram-Negative Bacteria.
Biochemistry, 57, 2018
|
|
6I38
 
 | |
