5X8M
 
 | | PD-L1 in complex with durvalumab | | 分子名称: | Programmed cell death 1 ligand 1, durvalumab heavy chain, durvalumab light chain | | 著者 | Heo, Y.S, Lee, H.T. | | 登録日 | 2017-03-03 | | 公開日 | 2017-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.661 Å) | | 主引用文献 | Molecular mechanism of PD-1/PD-L1 blockade via anti-PD-L1 antibodies atezolizumab and durvalumab
Sci Rep, 7, 2017
|
|
5WUX
 
 | | TNFalpha-certolizumab Fab | | 分子名称: | Tumor necrosis factor alpha, heavy, light | | 著者 | Heo, Y.S, Lee, J.U. | | 登録日 | 2016-12-21 | | 公開日 | 2017-06-07 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Molecular Basis for the Neutralization of Tumor Necrosis Factor alpha by Certolizumab Pegol in the Treatment of Inflammatory Autoimmune Diseases
Int J Mol Sci, 18, 2017
|
|
5X8L
 
 | | PD-L1 in complex with atezolizumab | | 分子名称: | Programmed cell death 1 ligand 1, atezolizumab heavy chain, atezolizumab light chain | | 著者 | Heo, Y.S, Lee, H.T. | | 登録日 | 2017-03-03 | | 公開日 | 2017-08-16 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Molecular mechanism of PD-1/PD-L1 blockade via anti-PD-L1 antibodies atezolizumab and durvalumab
Sci Rep, 7, 2017
|
|
3WE7
 
 | | Crystal Structure of Diacetylchitobiose Deacetylase from Pyrococcus horikoshii | | 分子名称: | ACETIC ACID, GLYCEROL, HEXANE-1,6-DIOL, ... | | 著者 | Mine, S, Nakamura, T, Fukuda, Y, Inoue, T, Uegaki, K, Sato, T. | | 登録日 | 2013-07-01 | | 公開日 | 2014-05-07 | | 最終更新日 | 2014-08-20 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Expression from engineered Escherichia coli chromosome and crystallographic study of archaeal N,N'-diacetylchitobiose deacetylase
Febs J., 281, 2014
|
|
2DUC
 
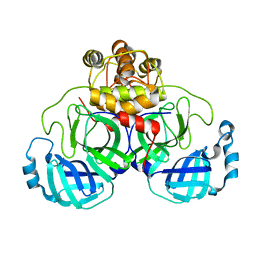 | | Crystal structure of SARS coronavirus main proteinase(3CLPRO) | | 分子名称: | Replicase polyprotein 1ab | | 著者 | Wang, H, Kim, Y.T, Muramatsu, T, Takemoto, C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-07-21 | | 公開日 | 2007-07-24 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | SARS-CoV 3CL protease cleaves its C-terminal autoprocessing site by novel subsite cooperativity
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
2E9B
 
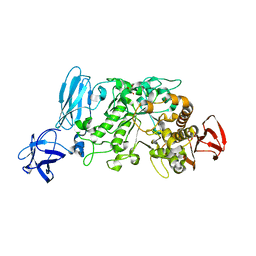 | | Crystal structure of pullulanase type I from Bacillus subtilis str. 168 complexed with maltose | | 分子名称: | ACETATE ION, AmyX protein, CALCIUM ION, ... | | 著者 | Mikami, B, Malle, D, Utsumi, S, Iwamoto, H, Katsuya, Y. | | 登録日 | 2007-01-24 | | 公開日 | 2008-02-19 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of pullulanase type I from Bacillus subtilis str. 168 in complex with maltose and alpha-cyclodextrin
To be Published
|
|
2E8Z
 
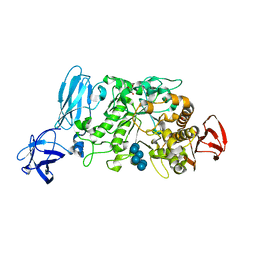 | | Crystal structure of pullulanase type I from Bacillus subtilis str. 168 complexed with alpha-cyclodextrin | | 分子名称: | ACETATE ION, AmyX protein, CALCIUM ION, ... | | 著者 | Mikami, B, Malle, D, Utsumi, S, Iwamoto, H, Katsuya, Y. | | 登録日 | 2007-01-24 | | 公開日 | 2008-02-19 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of pullulanase type I from Bacillus subtilis str. 168 in complex with maltose and alpha-cyclodextrin
To be Published
|
|
2E8Y
 
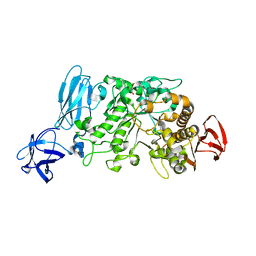 | | Crystal structure of pullulanase type I from Bacillus subtilis str. 168 | | 分子名称: | ACETATE ION, AmyX protein, CALCIUM ION, ... | | 著者 | Mikami, B, Malle, D, Utsumi, S, Iwamoto, H, Katsuya, Y. | | 登録日 | 2007-01-24 | | 公開日 | 2008-02-19 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Crystal structure of pullulanase type I from Bacillus subtilis str. 168
in complex with maltose and alpha-cyclodextrin
To be Published
|
|
2CDV
 
 | | REFINED STRUCTURE OF CYTOCHROME C3 AT 1.8 ANGSTROMS RESOLUTION | | 分子名称: | CYTOCHROME C3, HEME C | | 著者 | Higuchi, Y, Kusunoki, M, Matsuura, Y, Yasuoka, N, Kakudo, M. | | 登録日 | 1983-11-15 | | 公開日 | 1984-02-02 | | 最終更新日 | 2021-03-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Refined structure of cytochrome c3 at 1.8 A resolution
J.Mol.Biol., 172, 1984
|
|
