2G9G
 
 | | Crystal structure of His-tagged mouse PNGase C-terminal domain | | 分子名称: | GLYCEROL, SULFATE ION, peptide N-glycanase | | 著者 | Zhou, X, Zhao, G, Wang, L, Li, G, Lennarz, W.J, Schindelin, H. | | 登録日 | 2006-03-06 | | 公開日 | 2006-10-24 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and biochemical studies of the C-terminal domain of mouse peptide-N-glycanase identify it as a mannose-binding module.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2J9I
 
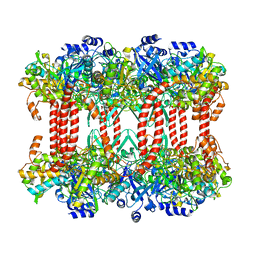 | | Lengsin is a survivor of an ancient family of class I glutamine synthetases in eukaryotes that has undergone evolutionary re- engineering for a tissue-specific role in the vertebrate eye lens. | | 分子名称: | GLUTAMATE-AMMONIA LIGASE DOMAIN-CONTAINING PROTEIN 1 | | 著者 | Wyatt, K, White, H.E, Wang, L, Bateman, O.A, Slingsby, C, Orlova, E.V, Wistow, G. | | 登録日 | 2006-11-09 | | 公開日 | 2006-12-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (17 Å) | | 主引用文献 | Lengsin is a Survivor of an Ancient Family of Class I Glutamine Synthetases Re-Engineered by Evolution for a Role in the Vertebrate Lens.
Structure, 14, 2006
|
|
2G9F
 
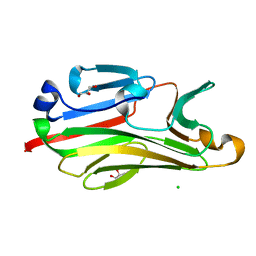 | | Crystal structure of intein-tagged mouse PNGase C-terminal domain | | 分子名称: | CHLORIDE ION, GLYCEROL, peptide N-glycanase | | 著者 | Zhou, X, Zhao, G, Wang, L, Li, G, Lennarz, W.J, Schindelin, H. | | 登録日 | 2006-03-06 | | 公開日 | 2006-10-24 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural and biochemical studies of the C-terminal domain of mouse peptide-N-glycanase identify it as a mannose-binding module.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HPJ
 
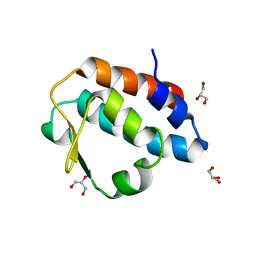 | | Crystal structure of the PUB domain of mouse PNGase | | 分子名称: | GLYCEROL, PNGase | | 著者 | Zhao, G, Zhou, X, Wang, L, Li, G, Lennarz, W, Schindelin, H. | | 登録日 | 2006-07-17 | | 公開日 | 2007-05-29 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Studies on peptide:N-glycanase-p97 interaction suggest that p97 phosphorylation modulates endoplasmic reticulum-associated degradation.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2I74
 
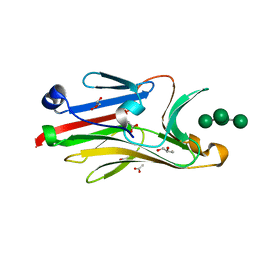 | | Crystal structure of mouse Peptide N-Glycanase C-terminal domain in complex with mannopentaose | | 分子名称: | ACETATE ION, GLYCEROL, PNGase, ... | | 著者 | Zhou, X, Zhao, G, Wang, L, Li, G, Lennarz, W.J, Schindelin, H. | | 登録日 | 2006-08-30 | | 公開日 | 2006-10-24 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural and biochemical studies of the C-terminal domain of mouse peptide-N-glycanase identify it as a mannose-binding module.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2JAT
 
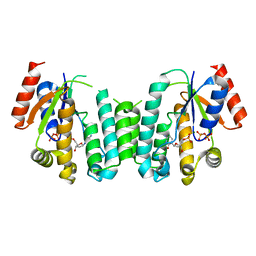 | | Structure of deoxyadenosine kinase from M.mycoides with products dcmp and a flexible dcdp bound | | 分子名称: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, DEOXYGUANOSINE KINASE, MAGNESIUM ION, ... | | 著者 | Welin, M, Wang, L, Eriksson, S, Eklund, H. | | 登録日 | 2006-11-30 | | 公開日 | 2007-01-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure-Function Analysis of a Bacterial Deoxyadenosine Kinase Reveals the Basis for Substrate Specificity.
J.Mol.Biol., 366, 2007
|
|
2JA1
 
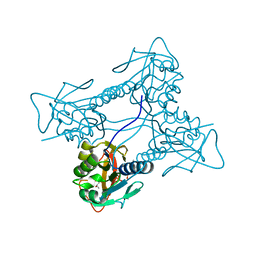 | | Thymidine kinase from B. cereus with TTP bound as phosphate donor. | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, THYMIDINE KINASE, THYMIDINE-5'-TRIPHOSPHATE, ... | | 著者 | Kosinska, U, Carnrot, C, Sandrini, M.P.B, Clausen, A.R, Wang, L, Piskur, J, Eriksson, S, Eklund, H. | | 登録日 | 2006-11-17 | | 公開日 | 2007-01-23 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural Studies of Thymidine Kinases from Bacillus Anthracis and Bacillus Cereus Provide Insights Into Quaternary Structure and Conformational Changes Upon Substrate Binding
FEBS J., 274, 2007
|
|
2JAS
 
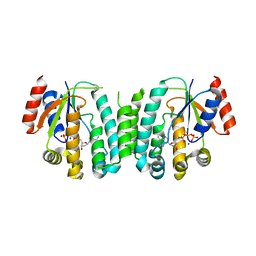 | | Structure of deoxyadenosine kinase from M.mycoides with bound dATP | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DEOXYGUANOSINE KINASE, MAGNESIUM ION | | 著者 | Welin, M, Wang, L, Eriksson, S, Eklund, H. | | 登録日 | 2006-11-30 | | 公開日 | 2007-01-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure-Function Analysis of a Bacterial Deoxyadenosine Kinase Reveals the Basis for Substrate Specificity.
J.Mol.Biol., 366, 2007
|
|
2J9R
 
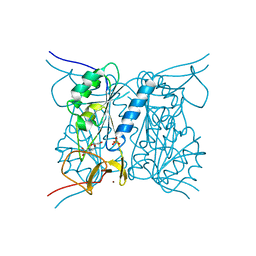 | | Thymidine kinase from B. anthracis in complex with dT. | | 分子名称: | PHOSPHATE ION, THYMIDINE, THYMIDINE KINASE, ... | | 著者 | Kosinska, U, Carnrot, C, Sandrini, M.P.B, Clausen, A.R, Wang, L, Piskur, J, Eriksson, S, Eklund, H. | | 登録日 | 2006-11-15 | | 公開日 | 2007-01-23 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural Studies of Thymidine Kinases from Bacillus Anthracis and Bacillus Cereus Provide Insights Into Quaternary Structure and Conformational Changes Upon Substrate Binding
FEBS J., 274, 2007
|
|
2JAQ
 
 | | Structure of deoxyadenosine kinase from M. mycoides with bound dCTP | | 分子名称: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DEOXYGUANOSINE KINASE | | 著者 | Welin, M, Wang, L, Eriksson, S, Eklund, H. | | 登録日 | 2006-11-30 | | 公開日 | 2007-01-03 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure-Function Analysis of a Bacterial Deoxyadenosine Kinase Reveals the Basis for Substrate Specificity.
J.Mol.Biol., 366, 2007
|
|
7WTK
 
 | | SARS-CoV-2 Omicron variant spike in complex with Fab XGv286 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of XGv286, ... | | 著者 | Wang, X, Fu, W. | | 登録日 | 2022-02-04 | | 公開日 | 2022-12-14 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
7WTG
 
 | | SARS-CoV-2 Omicron variant spike RBD in complex with Fab XGv051 | | 分子名称: | Heavy chain of XGv051, Light chain of XGv051, Spike protein S1 | | 著者 | Wang, X, Fu, W. | | 登録日 | 2022-02-04 | | 公開日 | 2022-12-14 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
7WTH
 
 | | SARS-CoV-2 Omicron variant spike RBD in complex with Fab XGv264 | | 分子名称: | Heavy chain of XGv264, Light chain of XGv264, Spike protein S1 | | 著者 | Wang, X, Fu, W. | | 登録日 | 2022-02-04 | | 公開日 | 2022-12-14 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
7WTJ
 
 | | SARS-CoV-2 Omicron variant spike RBD in complex with Fab XGv286 | | 分子名称: | Heavy chain of XGv286, Light chain of XGv286, Spike protein S1 | | 著者 | Wang, X, Fu, W. | | 登録日 | 2022-02-04 | | 公開日 | 2022-12-14 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
7WTI
 
 | | SARS-CoV-2 Omicron variant spike in complex with Fab XGv264 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of XGv264, Light chain of XGv264, ... | | 著者 | Wang, X, Fu, W. | | 登録日 | 2022-02-04 | | 公開日 | 2022-12-21 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
7WTF
 
 | | SARS-CoV-2 Omicron variant spike in complex with Fab XGv051 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of XGv051, ... | | 著者 | Wang, X, Fu, W. | | 登録日 | 2022-02-04 | | 公開日 | 2022-12-14 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
6EGE
 
 | |
6EGF
 
 | |
6EGD
 
 | |
8GQE
 
 | | Crystal structure of the W285A mutant of UVR8 in complex with RUP2 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ultraviolet-B receptor UVR8, WD repeat-containing protein RUP2 | | 著者 | Wang, Y.D, Wang, L.X, Guan, Z.Y, chang, H.F, Yin, P. | | 登録日 | 2022-08-30 | | 公開日 | 2022-09-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | RUP2 facilitates UVR8 redimerization via two interfaces.
Plant Commun., 4, 2023
|
|
8DZC
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor 17 | | 分子名称: | (3,5-difluorophenyl)methyl {(3S)-1-[(2S)-1-({(2S,3R)-4-(cyclopropylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-6-oxopiperidin-3-yl}carbamate, 3C-like proteinase nsp5, GLYCEROL | | 著者 | Sacco, M, Chen, Y. | | 登録日 | 2022-08-06 | | 公開日 | 2023-04-05 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Development of the Safe and Broad-Spectrum Aldehyde and Ketoamide Mpro inhibitors Derived from the Constrained alpha , gamma-AA Peptide Scaffold.
Chemistry, 29, 2023
|
|
8DZB
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor 11 | | 分子名称: | 3C-like proteinase nsp5, GLYCEROL, benzyl {(3S)-1-[(2S)-1-({(2S,3R)-4-(cyclopropylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-5-oxopyrrolidin-3-yl}carbamate | | 著者 | Sacco, M, Chen, Y. | | 登録日 | 2022-08-06 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Development of the Safe and Broad-Spectrum Aldehyde and Ketoamide Mpro inhibitors Derived from the Constrained alpha , gamma-AA Peptide Scaffold.
Chemistry, 29, 2023
|
|
4C13
 
 | | x-ray crystal structure of Staphylococcus aureus MurE with UDP-MurNAc- Ala-Glu-Lys | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Ruane, K.M, Roper, D.I, Fulop, V, Barreteau, H, Boniface, A, Dementin, S, Blanot, D, Mengin-Lecreulx, D, Gobec, S, Dessen, A, Dowson, C.G, Lloyd, A.J. | | 登録日 | 2013-08-09 | | 公開日 | 2013-10-02 | | 最終更新日 | 2021-03-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Discovery of a first-in-class CDK2 selective degrader for AML differentiation therapy.
Nat.Chem.Biol., 2021
|
|
5UEX
 
 | | BRD4_BD2_A-1497627 | | 分子名称: | 17-{[ethyl(dihydroxy)-lambda~4~-sulfanyl]amino}-11,13-difluoro-2-methyl-6,7,8,9-tetrahydrodibenzo[4,5:7,8][1,6]dioxacyclododecino[3,2-c]pyridin-3(2H)-one, Bromodomain-containing protein 4 | | 著者 | Park, C.H. | | 登録日 | 2017-01-03 | | 公開日 | 2017-05-10 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Fragment-Based, Structure-Enabled Discovery of Novel Pyridones and Pyridone Macrocycles as Potent Bromodomain and Extra-Terminal Domain (BET) Family Bromodomain Inhibitors.
J. Med. Chem., 60, 2017
|
|
5UEU
 
 | | BRD4_BD2_A-1107604 | | 分子名称: | Bromodomain-containing protein 4, methyl [(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]acetate | | 著者 | Park, C.H. | | 登録日 | 2017-01-03 | | 公開日 | 2017-05-10 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Fragment-Based, Structure-Enabled Discovery of Novel Pyridones and Pyridone Macrocycles as Potent Bromodomain and Extra-Terminal Domain (BET) Family Bromodomain Inhibitors.
J. Med. Chem., 60, 2017
|
|
