8SV6
 
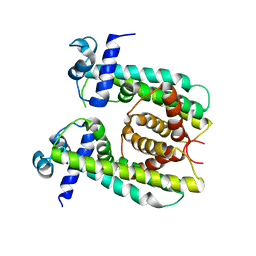 | | Structure of the M. smegmatis DarR protein | | 分子名称: | Fatty acid metabolism regulator protein | | 著者 | Schumacher, M.A. | | 登録日 | 2023-05-15 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.56 Å) | | 主引用文献 | Structures of the DarR transcription regulator reveal unique modes of second messenger and DNA binding.
Nat Commun, 14, 2023
|
|
8SVA
 
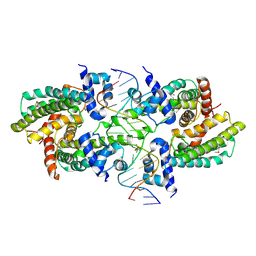 | | Structure of the Rhodococcus sp. USK13 DarR-20 bp DNA complex | | 分子名称: | DNA (5'-D(*TP*AP*GP*AP*TP*AP*CP*TP*CP*CP*GP*GP*AP*GP*TP*AP*TP*CP*TP*A)-3'), PHOSPHATE ION, TetR/AcrR family transcriptional regulator | | 著者 | Schumacher, M.A. | | 登録日 | 2023-05-15 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.96 Å) | | 主引用文献 | Structures of the DarR transcription regulator reveal unique modes of second messenger and DNA binding.
Nat Commun, 14, 2023
|
|
8SUA
 
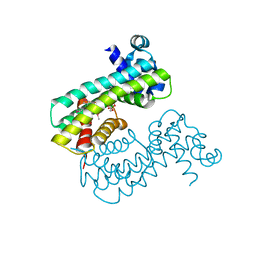 | | Structure of M. baixiangningiae DarR-ligand complex | | 分子名称: | 3-azanyl-3-(hydroxymethyl)-1,5,7,11-tetraoxa-6$l^{4}-boraspiro[5.5]undecan-9-ol, DarR | | 著者 | Schumacher, M.A. | | 登録日 | 2023-05-11 | | 公開日 | 2023-11-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structures of the DarR transcription regulator reveal unique modes of second messenger and DNA binding.
Nat Commun, 14, 2023
|
|
8SVD
 
 | |
8TFK
 
 | |
8TFC
 
 | |
8TFB
 
 | |
8TGE
 
 | |
8UFJ
 
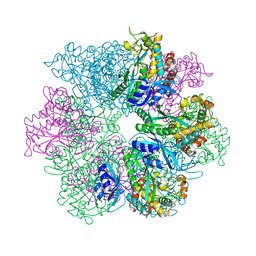 | | Structure of M. mazei GS(R167L-A168G) apo form | | 分子名称: | Glutamine synthetase, MAGNESIUM ION | | 著者 | Schumacher, M.A. | | 登録日 | 2023-10-04 | | 公開日 | 2023-11-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | M. mazei glutamine synthetase and glutamine synthetase-GlnK1 structures reveal enzyme regulation by oligomer modulation.
Nat Commun, 14, 2023
|
|
8TP8
 
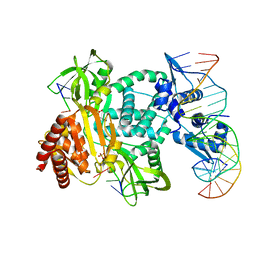 | | Structure of the C. crescentus WYL-activator, DriD, bound to ssDNA and cognate DNA | | 分子名称: | DNA (5'-D(*AP*TP*AP*CP*GP*AP*CP*AP*GP*TP*AP*AP*CP*TP*GP*TP*CP*GP*TP*AP*T)-3'), DNA (5'-D(*AP*TP*AP*CP*GP*AP*CP*AP*GP*TP*TP*AP*CP*TP*GP*TP*CP*GP*TP*AP*T)-3'), DNA (5'-D(P*GP*TP*C)-3'), ... | | 著者 | Schumacher, M.A. | | 登録日 | 2023-08-04 | | 公開日 | 2023-11-29 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.74 Å) | | 主引用文献 | Structure of the WYL-domain containing transcription activator, DriD, in complex with ssDNA effector and DNA target site.
Nucleic Acids Res., 52, 2024
|
|
8TPK
 
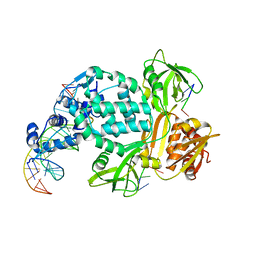 | | P6522 crystal form of C. crescentus DriD-ssDNA-DNA complex | | 分子名称: | DNA (5'-D(*AP*TP*AP*CP*GP*AP*CP*AP*GP*TP*AP*AP*CP*TP*GP*TP*CP*GP*TP*AP*T)-3'), DNA (5'-D(*AP*TP*AP*CP*GP*AP*CP*AP*GP*TP*TP*AP*CP*TP*GP*TP*CP*GP*TP*AP*T)-3'), DNA (5'-D(P*GP*TP*C)-3'), ... | | 著者 | Schumacher, M.A. | | 登録日 | 2023-08-04 | | 公開日 | 2023-11-29 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.46 Å) | | 主引用文献 | Structure of the WYL-domain containing transcription activator, DriD, in complex with ssDNA effector and DNA target site.
Nucleic Acids Res., 52, 2024
|
|
1HAB
 
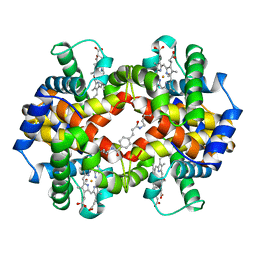 | | CROSSLINKED HAEMOGLOBIN | | 分子名称: | 4-CARBOXYCINNAMIC ACID, CARBON MONOXIDE, HEMOGLOBIN A, ... | | 著者 | Schumacher, M.A, Dixon, M.M, Kluger, R, Jones, R.T, Brennan, R.G. | | 登録日 | 1996-03-13 | | 公開日 | 1997-11-12 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Allosteric intermediates indicate R2 is the liganded hemoglobin end state.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1ZX4
 
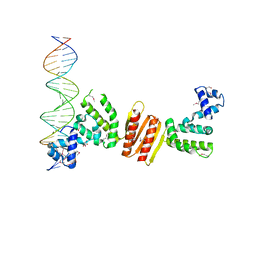 | | Structure of ParB bound to DNA | | 分子名称: | CITRIC ACID, Plasmid Partition par B protein, parS-small DNA centromere site | | 著者 | Schumacher, M.A, Funnell, B.E. | | 登録日 | 2005-06-06 | | 公開日 | 2005-11-29 | | 最終更新日 | 2017-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | Structures of ParB bound to DNA reveal mechanism of partition complex formation.
Nature, 438, 2005
|
|
1HAC
 
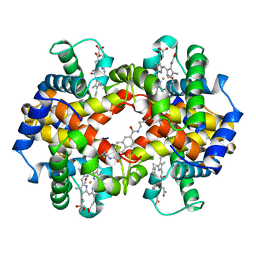 | | CROSSLINKED HAEMOGLOBIN | | 分子名称: | 2,6-DICARBOXYNAPHTHALENE, CARBON MONOXIDE, HEMOGLOBIN A, ... | | 著者 | Schumacher, M.A, Dixon, M.M, Kluger, R, Jones, R.T, Brennan, R.G. | | 登録日 | 1996-03-13 | | 公開日 | 1997-11-12 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Allosteric intermediates indicate R2 is the liganded hemoglobin end state.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
2NTZ
 
 | |
2NZV
 
 | | Structural mechanism for the fine-tuning of CcpA function by the small molecule effectors G6P and FBP | | 分子名称: | 1,6-di-O-phosphono-beta-D-fructofuranose, Catabolite control protein, Phosphocarrier protein HPr, ... | | 著者 | Schumacher, M.A, Hillen, W, Brennan, R.G. | | 登録日 | 2006-11-25 | | 公開日 | 2007-05-01 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural Mechanism for the Fine-tuning of CcpA Function by The Small Molecule Effectors Glucose 6-Phosphate and Fructose 1,6-Bisphosphate.
J.Mol.Biol., 368, 2007
|
|
2NZU
 
 | | Structural mechanism for the fine-tuning of CcpA function by the small molecule effectors G6P and FBP | | 分子名称: | 6-O-phosphono-beta-D-glucopyranose, Catabolite control protein, Phosphocarrier protein HPr, ... | | 著者 | Schumacher, M.A, Hillen, W, Brennan, R.G. | | 登録日 | 2006-11-25 | | 公開日 | 2007-05-01 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural Mechanism for the Fine-tuning of CcpA Function by The Small Molecule Effectors Glucose 6-Phosphate and Fructose 1,6-Bisphosphate.
J.Mol.Biol., 368, 2007
|
|
2OEN
 
 | | Structural mechanism for the fine-tuning of CcpA function by the small molecule effectors glucose-6-phosphate and fructose-1,6-bisphosphate | | 分子名称: | Catabolite control protein, Phosphocarrier protein HPr | | 著者 | Schumacher, M.A, Seidel, G, Hillen, W, Brennan, R.G. | | 登録日 | 2006-12-30 | | 公開日 | 2007-05-01 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3.17 Å) | | 主引用文献 | Structural Mechanism for the Fine-tuning of CcpA Function by The Small Molecule Effectors Glucose 6-Phosphate and Fructose 1,6-Bisphosphate.
J.Mol.Biol., 368, 2007
|
|
1ZVV
 
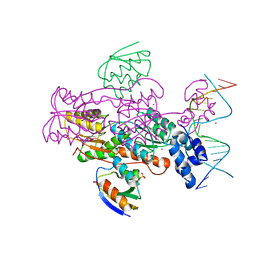 | | Crystal structure of a ccpa-crh-dna complex | | 分子名称: | DNA recognition strand CRE, Glucose-resistance amylase regulator, HPr-like protein crh, ... | | 著者 | Schumacher, M.A, Brennan, R.G, Hillen, W, Seidel, G. | | 登録日 | 2005-06-02 | | 公開日 | 2006-02-07 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | Phosphoprotein Crh-Ser46-P displays altered binding to CcpA to effect carbon catabolite regulation.
J.Biol.Chem., 281, 2006
|
|
4YG1
 
 | |
4YG4
 
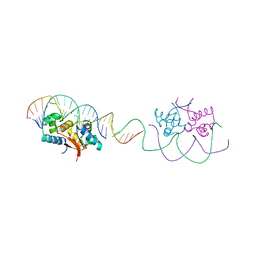 | | HipB-O1-O1* complex | | 分子名称: | Antitoxin HipB, DNA (28-MER), DNA (5'-D(*AP*TP*AP*TP*CP*CP*CP*CP*TP*TP*AP*AP*GP*GP*GP*GP*AP*TP*AP*A)-3') | | 著者 | Schumacher, M.A. | | 登録日 | 2015-02-25 | | 公開日 | 2015-07-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | HipBA-promoter structures reveal the basis of heritable multidrug tolerance.
Nature, 524, 2015
|
|
4YG7
 
 | |
6AMK
 
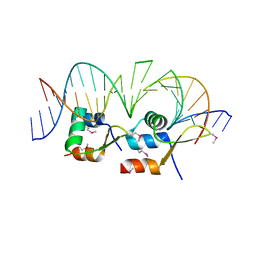 | | Structure of Streptomyces venezuelae BldC-whiI opt complex | | 分子名称: | DNA (5'-D(*AP*AP*TP*GP*TP*CP*CP*GP*AP*AP*TP*TP*AP*CP*CP*CP*GP*AP*AP*TP*TP*G)-3'), DNA (5'-D(*TP*TP*CP*AP*AP*TP*TP*CP*GP*GP*GP*TP*AP*AP*TP*TP*CP*GP*GP*GP*CP*A)-3'), Putative DNA-binding protein | | 著者 | Schumacher, M.A. | | 登録日 | 2017-08-09 | | 公開日 | 2018-03-28 | | 最終更新日 | 2018-11-07 | | 実験手法 | X-RAY DIFFRACTION (3.288 Å) | | 主引用文献 | The MerR-like protein BldC binds DNA direct repeats as cooperative multimers to regulate Streptomyces development.
Nat Commun, 9, 2018
|
|
6AMA
 
 | |
4PTA
 
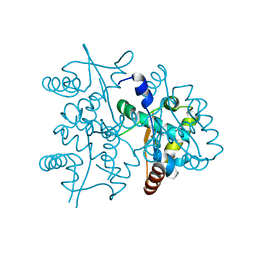 | | Structure of MDR initiator | | 分子名称: | Replication initiator protein | | 著者 | Schumacher, M.A. | | 登録日 | 2014-03-10 | | 公開日 | 2014-06-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.6003 Å) | | 主引用文献 | Mechanism of staphylococcal multiresistance plasmid replication origin assembly by the RepA protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
