4TJV
 
 | | Crystal structure of protease-associated domain of Arabidopsis vacuolar sorting receptor 1 | | 分子名称: | IODIDE ION, Vacuolar-sorting receptor 1 | | 著者 | Luo, F, Fong, Y.H, Jiang, L.W, Wong, K.B. | | 登録日 | 2014-05-25 | | 公開日 | 2014-12-10 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.651 Å) | | 主引用文献 | How vacuolar sorting receptor proteins interact with their cargo proteins: crystal structures of apo and cargo-bound forms of the protease-associated domain from an Arabidopsis vacuolar sorting receptor.
Plant Cell, 26, 2014
|
|
7O0W
 
 | | Cryo-EM structure of the RC-dLH complex (model_1b) from Gemmatimonas phototrophica at 2.47 A | | 分子名称: | (19R,22S)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9Z)-hexadec-9-enoate, (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, ... | | 著者 | Qian, P, Koblizek, M. | | 登録日 | 2021-03-27 | | 公開日 | 2022-03-02 | | 最終更新日 | 2022-03-16 | | 実験手法 | ELECTRON MICROSCOPY (2.47 Å) | | 主引用文献 | 2.4- angstrom structure of the double-ring Gemmatimonas phototrophica photosystem.
Sci Adv, 8, 2022
|
|
7O0U
 
 | | Cryo-EM structure (model_1a) of the RC-dLH complex from Gemmatimonas phototrophica at 2.4 A | | 分子名称: | (19R,22S)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9Z)-hexadec-9-enoate, (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, ... | | 著者 | Qian, P, Koblizek, M. | | 登録日 | 2021-03-27 | | 公開日 | 2022-03-02 | | 最終更新日 | 2022-03-16 | | 実験手法 | ELECTRON MICROSCOPY (2.35 Å) | | 主引用文献 | 2.4- angstrom structure of the double-ring Gemmatimonas phototrophica photosystem.
Sci Adv, 8, 2022
|
|
7O0V
 
 | | Cryo-EM structure (model_2a) of the RC-dLH complex from Gemmatimonas phototrophica at 2.5 A | | 分子名称: | (19R,22S)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9Z)-hexadec-9-enoate, (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, ... | | 著者 | Qian, P, Koblizek, M. | | 登録日 | 2021-03-27 | | 公開日 | 2022-03-02 | | 最終更新日 | 2022-03-16 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | 2.4- angstrom structure of the double-ring Gemmatimonas phototrophica photosystem.
Sci Adv, 8, 2022
|
|
7O0X
 
 | | Cryo-EM structure (model_2b) of the RC-dLH complex from Gemmatimonas phototrophica at 2.44 A | | 分子名称: | (19R,22S)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9Z)-hexadec-9-enoate, (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, ... | | 著者 | Qian, P, Koblizek, M. | | 登録日 | 2021-03-28 | | 公開日 | 2022-03-02 | | 最終更新日 | 2022-03-16 | | 実験手法 | ELECTRON MICROSCOPY (2.44 Å) | | 主引用文献 | 2.4- angstrom structure of the double-ring Gemmatimonas phototrophica photosystem.
Sci Adv, 8, 2022
|
|
8WVD
 
 | | Crystal structure of Glycosyltransferase in complex with UD1 | | 分子名称: | Glycosyltransferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | 著者 | Li, J, Shan, N, Yang, J.G, Liu, W.D, Sun, Y.X. | | 登録日 | 2023-10-23 | | 公開日 | 2024-09-04 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Enzymatic Synthesis of Novel Terpenoid Glycoside Derivatives Decorated with N -Acetylglucosamine Catalyzed by UGT74AC1.
J.Agric.Food Chem., 72, 2024
|
|
8KCB
 
 | |
8KCC
 
 | |
7YJ0
 
 | |
8WY7
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor 22 | | 分子名称: | 2-[[5-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-1-methyl-2-oxidanylidene-pyridin-4-yl]amino]-~{N}-(4-oxidanylcyclohexyl)ethanamide, Bromodomain-containing protein 4 | | 著者 | Xu, H, Zhao, X, Shen, H, Xu, Y, Wu, X. | | 登録日 | 2023-10-30 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.83 Å) | | 主引用文献 | Discovery of Novel Phenoxyaryl Pyridones as Bromodomain and Extra-Terminal Domain (BET) Inhibitors with High Selectivity for the Second Bromodomain (BD2) to Potentially Treat Acute Myeloid Leukemia.
J.Med.Chem., 67, 2024
|
|
8WY3
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor 21 | | 分子名称: | Bromodomain-containing protein 4, ~{N}-cyclopropyl-2-[[5-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-1-methyl-2-oxidanylidene-pyridin-4-yl]amino]ethanamide | | 著者 | Xu, H, Zhao, X, Shen, H, Xu, Y, Wu, X. | | 登録日 | 2023-10-30 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.78 Å) | | 主引用文献 | Discovery of Novel Phenoxyaryl Pyridones as Bromodomain and Extra-Terminal Domain (BET) Inhibitors with High Selectivity for the Second Bromodomain (BD2) to Potentially Treat Acute Myeloid Leukemia.
J.Med.Chem., 67, 2024
|
|
8WYG
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor 22 | | 分子名称: | 2-[[5-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-1-methyl-2-oxidanylidene-pyridin-4-yl]amino]-~{N}-(4-oxidanylcyclohexyl)ethanamide, Bromodomain-containing protein 2 | | 著者 | Xu, H, Zhao, X, Shen, H, Xu, Y, Wu, X. | | 登録日 | 2023-10-30 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3.13 Å) | | 主引用文献 | Discovery of Novel Phenoxyaryl Pyridones as Bromodomain and Extra-Terminal Domain (BET) Inhibitors with High Selectivity for the Second Bromodomain (BD2) to Potentially Treat Acute Myeloid Leukemia.
J.Med.Chem., 67, 2024
|
|
8WXY
 
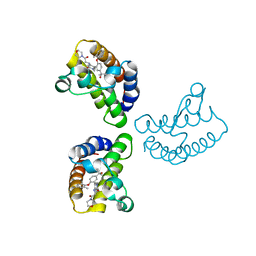 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor 23 | | 分子名称: | 5-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-1-methyl-4-[(2-morpholin-4-yl-2-oxidanylidene-ethyl)amino]pyridin-2-one, Bromodomain-containing protein 4, GLYCEROL | | 著者 | Xu, H, Zhao, X, Shen, H, Xu, Y, Wu, X. | | 登録日 | 2023-10-30 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.87 Å) | | 主引用文献 | Discovery of Novel Phenoxyaryl Pyridones as Bromodomain and Extra-Terminal Domain (BET) Inhibitors with High Selectivity for the Second Bromodomain (BD2) to Potentially Treat Acute Myeloid Leukemia.
J.Med.Chem., 67, 2024
|
|
7YP9
 
 | |
7YPA
 
 | |
7YPB
 
 | |
3K45
 
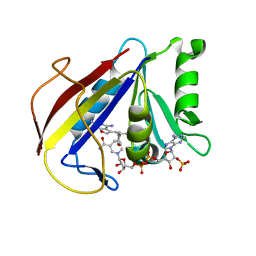 | | Alternate Binding Modes Observed for the E- and Z-isomers of 2,4-Diaminofuro[2,3d]pyrimidines as Ternary Complexes with NADPH and Mouse Dihydrofolate Reductase | | 分子名称: | 5-[(1Z)-2-(2-methoxyphenyl)prop-1-en-1-yl]furo[2,3-d]pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Cody, V. | | 登録日 | 2009-10-05 | | 公開日 | 2009-10-13 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Design, synthesis, and X-ray crystal structures of 2,4-diaminofuro[2,3-d]pyrimidines as multireceptor tyrosine kinase and dihydrofolate reductase inhibitors.
Bioorg.Med.Chem., 17, 2009
|
|
3K47
 
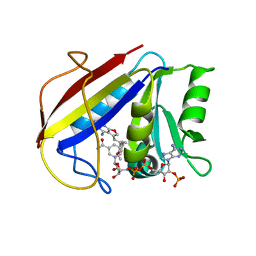 | | Alternate Binding Modes Observed for the E- and Z-Isomers of 2,4-Diaminofuro[2,3-d]pyrimidines as Ternary Complexes with NADPH and Mouse Dihydrofolate Reductase | | 分子名称: | 5-[(1E)-2-(2-methoxyphenyl)prop-1-en-1-yl]furo[2,3-d]pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Cody, V, Pace, J, Queener, S.F, Gangjee, A. | | 登録日 | 2009-10-05 | | 公開日 | 2009-10-13 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Design, synthesis, and X-ray crystal structures of 2,4-diaminofuro[2,3-d]pyrimidines as multireceptor tyrosine kinase and dihydrofolate reductase inhibitors.
Bioorg.Med.Chem., 17, 2009
|
|
6ML6
 
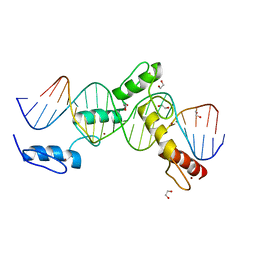 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 4 with a CpA 5mC Modification) | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*CP*GP*(5CM)P*AP*GP*GP*TP*CP*CP*TP*GP*GP*AP*CP*GP*AP*AP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*TP*TP*CP*GP*TP*CP*CP*AP*GP*GP*AP*CP*CP*TP*GP*CP*G)-3'), ... | | 著者 | Horton, J.R, Cheng, X, Ren, R. | | 登録日 | 2018-09-26 | | 公開日 | 2019-07-03 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
6ML5
 
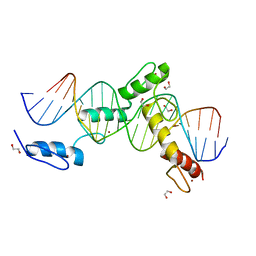 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 4) | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*CP*GP*CP*AP*GP*GP*TP*CP*CP*TP*GP*GP*AP*CP*GP*AP*AP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*TP*TP*CP*GP*TP*CP*CP*AP*GP*GP*AP*CP*CP*TP*GP*CP*G)-3'), ... | | 著者 | Horton, J.R, Cheng, X, Ren, R. | | 登録日 | 2018-09-26 | | 公開日 | 2019-07-03 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
6ML2
 
 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 1) | | 分子名称: | DNA (5'-D(*AP*CP*GP*CP*AP*GP*GP*TP*CP*CP*TP*GP*GP*CP*AP*GP*CP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP*AP*CP*CP*TP*GP*CP*G)-3'), ZINC ION, ... | | 著者 | Horton, J.R, Cheng, X, Ren, R. | | 登録日 | 2018-09-26 | | 公開日 | 2019-07-03 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.874 Å) | | 主引用文献 | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
6ML7
 
 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 4 with a CpG 5mC Modification) | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*AP*CP*GP*CP*AP*GP*GP*TP*CP*CP*TP*GP*GP*AP*(5CM)P*GP*AP*AP*TP*T)-3'), ... | | 著者 | Horton, J.R, Cheng, X, Ren, R. | | 登録日 | 2018-09-26 | | 公開日 | 2019-07-03 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
6ML3
 
 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 2) | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*CP*GP*CP*AP*GP*GP*TP*CP*CP*TP*GP*GP*AP*AP*GP*CP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*GP*CP*TP*TP*CP*CP*AP*GP*GP*AP*CP*CP*TP*GP*CP*G)-3'), ... | | 著者 | Horton, J.R, Cheng, X, Ren, R. | | 登録日 | 2018-09-26 | | 公開日 | 2019-07-03 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.683 Å) | | 主引用文献 | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
6ML4
 
 | | BTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 3) | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*CP*GP*CP*AP*GP*GP*TP*CP*CP*TP*GP*GP*AP*CP*GP*AP*AP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*TP*TP*CP*GP*TP*CP*CP*AP*GP*GP*AP*CP*CP*TP*GP*CP*G)-3'), ... | | 著者 | Horton, J.R, Cheng, X, Ren, R. | | 登録日 | 2018-09-26 | | 公開日 | 2019-07-03 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.482 Å) | | 主引用文献 | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
5WUE
 
 | | Structural basis for conductance through TRIC cation channels | | 分子名称: | SULFATE ION, Uncharacterized protein | | 著者 | Su, M, Gao, F, Mao, Y, Li, D.L, Guo, Y.Z, Wang, X.H, Bruni, R, Kloss, B, Hendrickson, W.A, Chen, Y.H, New York Consortium on Membrane Protein Structure (NYCOMPS) | | 登録日 | 2016-12-17 | | 公開日 | 2017-06-21 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis for conductance through TRIC cation channels.
Nat Commun, 8, 2017
|
|
