7CJ5
 
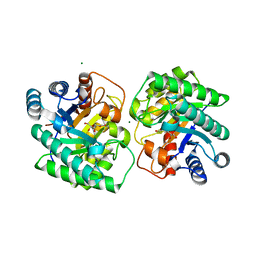 | | Crystal structure of homo dimeric D-allulose 3-epimerase from Methylomonas sp. in complex with D-fructose | | 分子名称: | D-fructose, Epimerase, MAGNESIUM ION, ... | | 著者 | Yoshida, H, Yoshihara, A, Kamitori, S. | | 登録日 | 2020-07-09 | | 公開日 | 2021-04-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of a novel homodimeric l-ribulose 3-epimerase from Methylomonus sp.
Febs Open Bio, 11, 2021
|
|
7VQP
 
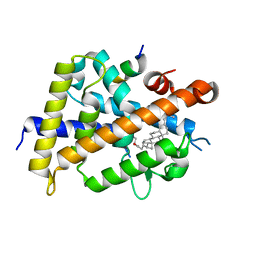 | | Vitamin D receptor complexed with a lithocholic acid derivative | | 分子名称: | 3-((R)-4-((3R,5R,8R,9S,10S,13R,14S,17R)-3-(2-hydroxy-2-methylpropyl)-10,13-dimethylhexadecahydro-1H-cyclopenta[a]phenanthren-17-yl)pentanamido)propanoic acid, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | 著者 | Kato, K, Numoto, N, Kagechika, H, Tanatani, A, Ito, N. | | 登録日 | 2021-10-20 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Lithocholic Acid Amides as Potent Vitamin D Receptor Agonists.
Biomolecules, 12, 2022
|
|
4GJJ
 
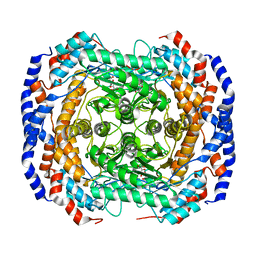 | |
4GJI
 
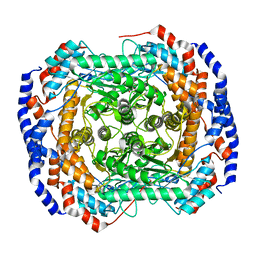 | |
7C7V
 
 | | Vitamin D3 receptor/lithochoric acid derivative complex | | 分子名称: | (4R)-4-[(3R,5R,8R,9S,10S,13R,14S,17R)-10,13-dimethyl-3-(2-methyl-2-oxidanyl-propyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]pentanoic acid, FORMIC ACID, Mediator of RNA polymerase II transcription subunit 1, ... | | 著者 | Masuno, H, Numoto, N, Kagechika, H, Ito, N. | | 登録日 | 2020-05-26 | | 公開日 | 2021-01-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Lithocholic Acid Derivatives as Potent Vitamin D Receptor Agonists.
J.Med.Chem., 64, 2021
|
|
7C7W
 
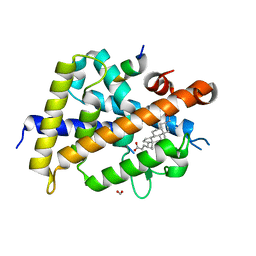 | | Vitamin D3 receptor/lithochoric acid derivative complex | | 分子名称: | (4R)-4-[(3S,5R,8R,9S,10S,13R,14S,17R)-10,13-dimethyl-3-(2-methyl-2-oxidanyl-propyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]pentanoic acid, FORMIC ACID, Mediator of RNA polymerase II transcription subunit 1, ... | | 著者 | Masuno, H, Numoto, N, Kagechika, H, Ito, N. | | 登録日 | 2020-05-26 | | 公開日 | 2021-01-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Lithocholic Acid Derivatives as Potent Vitamin D Receptor Agonists.
J.Med.Chem., 64, 2021
|
|
3VYL
 
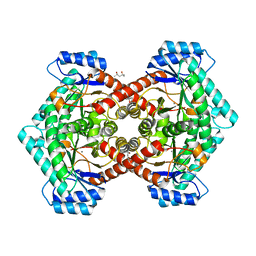 | | Structure of L-ribulose 3-epimerase | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, L-ribulose 3-epimerase, MANGANESE (II) ION | | 著者 | Uechi, K, Sakuraba, H, Takata, G. | | 登録日 | 2012-09-27 | | 公開日 | 2013-10-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural insight into L-ribulose 3-epimerase from Mesorhizobium loti.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
