8IFF
 
 | | Cryo-EM structure of Arabidopsis phytochrome A. | | 分子名称: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome A | | 著者 | Ma, L, Zhou, C, Wang, J, Guan, Z, Yin, P. | | 登録日 | 2023-02-17 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Plant phytochrome A in the Pr state assembles as an asymmetric dimer.
Cell Res., 33, 2023
|
|
4GJP
 
 | | Crystal structure of the TAL effector dHax3 bound to dsDNA containing repetitive methyl-CpG | | 分子名称: | DNA (5'-D(*AP*GP*AP*GP*AP*CP*GP*CP*GP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*(5CM)P*GP*(5CM)P*GP*TP*CP*TP*CP*T)-3'), Hax3, ... | | 著者 | Yan, N, Deng, D, Yan, C.Y, Yin, P, Pan, X.J, Shi, Y.G. | | 登録日 | 2012-08-10 | | 公開日 | 2012-10-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Crystal structure of a protein complex
To be Published
|
|
4GJR
 
 | | Crystal structure of the TAL effector dHax3 bound to methylated dsDNA | | 分子名称: | DNA (5'-D(*AP*GP*GP*GP*AP*GP*GP*TP*AP*GP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*(5CM)P*TP*AP*(5CM)P*CP*TP*CP*(5CM)P*CP*T)-3'), Hax3 | | 著者 | Yan, N, Deng, D, Yan, C.Y, Yin, P, Pan, X.J, Shi, Y.G. | | 登録日 | 2012-08-10 | | 公開日 | 2012-10-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structure of a protein complex
To be Published
|
|
7R6N
 
 | | Exon-free state of the Tetrahymena group I intron, symmetry-expanded monomer from a synthetic trimeric construct | | 分子名称: | Group I intron, MAGNESIUM ION | | 著者 | Thelot, F, Liu, D, Liao, M, Yin, P. | | 登録日 | 2021-06-22 | | 公開日 | 2022-05-04 | | 最終更新日 | 2022-06-01 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Sub-3- angstrom cryo-EM structure of RNA enabled by engineered homomeric self-assembly.
Nat.Methods, 19, 2022
|
|
7R6M
 
 | | Post-2S intermediate of the Tetrahymena group I intron, symmetry-expanded monomer from a synthetic dimeric construct | | 分子名称: | Group I intron, Ligated exon mimic of the Group I intron, MAGNESIUM ION | | 著者 | Thelot, F, Liu, D, Liao, M, Yin, P. | | 登録日 | 2021-06-22 | | 公開日 | 2022-05-04 | | 最終更新日 | 2022-06-01 | | 実験手法 | ELECTRON MICROSCOPY (3.68 Å) | | 主引用文献 | Sub-3- angstrom cryo-EM structure of RNA enabled by engineered homomeric self-assembly.
Nat.Methods, 19, 2022
|
|
7R6L
 
 | | 5 prime exon-free pre-2S intermediate of the Tetrahymena group I intron, symmetry-expanded monomer from a synthetic dimeric construct | | 分子名称: | Group I intron, 3 prime fragment plus 3 prime exon, 5 prime fragment, ... | | 著者 | Thelot, F, Liu, D, Liao, M, Yin, P. | | 登録日 | 2021-06-22 | | 公開日 | 2022-05-04 | | 最終更新日 | 2022-06-01 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Sub-3- angstrom cryo-EM structure of RNA enabled by engineered homomeric self-assembly.
Nat.Methods, 19, 2022
|
|
8HAV
 
 | | An auto-activation mechanism of plant non-specific phospholipase C | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Non-specific phospholipase C4 | | 著者 | Zhao, F, Fan, R.Y, Guan, Z.Y, Guo, L, Yin, P. | | 登録日 | 2022-10-26 | | 公開日 | 2023-01-25 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Insights into the mechanism of phospholipid hydrolysis by plant non-specific phospholipase C.
Nat Commun, 14, 2023
|
|
8HAW
 
 | | An auto-activation mechanism of plant non-specific phospholipase C | | 分子名称: | CALCIUM ION, GLYCEROL, Non-specific phospholipase C4, ... | | 著者 | Zhao, F, Fan, R.Y, Guan, Z.Y, Guo, L, Yin, P. | | 登録日 | 2022-10-26 | | 公開日 | 2023-01-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Insights into the mechanism of phospholipid hydrolysis by plant non-specific phospholipase C.
Nat Commun, 14, 2023
|
|
5WWD
 
 | | Crystal structure of AtNUDX1 | | 分子名称: | AMMONIUM ION, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Liu, J, Guan, Z, Yan, L, Zou, T, Yin, P. | | 登録日 | 2016-12-31 | | 公開日 | 2017-11-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.386 Å) | | 主引用文献 | Structural Insights into the Substrate Recognition Mechanism of Arabidopsis GPP-Bound NUDX1 for Noncanonical Monoterpene Biosynthesis.
Mol Plant, 11, 2018
|
|
5WY6
 
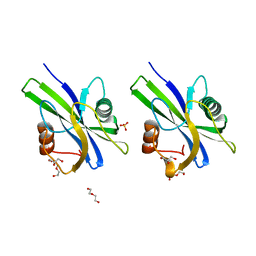 | | Crystal structure of AtNUDX1 (E56A) | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Nudix hydrolase 1, ... | | 著者 | Liu, J, Guan, Z, Yan, L, Zou, T, Yin, P. | | 登録日 | 2017-01-11 | | 公開日 | 2017-11-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.779 Å) | | 主引用文献 | Structural Insights into the Substrate Recognition Mechanism of Arabidopsis GPP-Bound NUDX1 for Noncanonical Monoterpene Biosynthesis.
Mol Plant, 11, 2018
|
|
5XYB
 
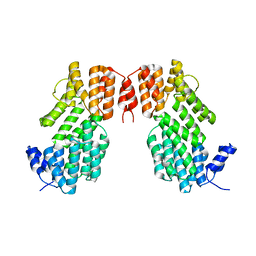 | |
5Y24
 
 | | Crystal structure of AimR from Bacillus phage SPbeta in complex with its signalling peptide | | 分子名称: | AimR transcriptional regulator, BROMIDE ION, GLY-MET-PRO-ARG-GLY-ALA | | 著者 | Wang, Q, Guan, Z.Y, Zou, T.T, Yin, P. | | 登録日 | 2017-07-24 | | 公開日 | 2018-09-19 | | 最終更新日 | 2018-11-28 | | 実験手法 | X-RAY DIFFRACTION (1.922 Å) | | 主引用文献 | Structural basis of the arbitrium peptide-AimR communication system in the phage lysis-lysogeny decision.
Nat Microbiol, 3, 2018
|
|
5YGU
 
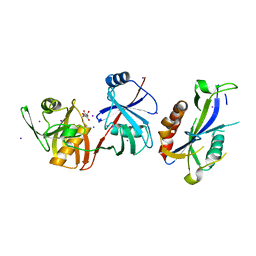 | | Crystal structure of Escherichia coli (strain K12) mRNA Decapping Complex RppH-DapF | | 分子名称: | Diaminopimelate epimerase, IODIDE ION, L(+)-TARTARIC ACID, ... | | 著者 | Wang, Q, Guan, Z.Y, Zhang, D.L, Zou, T.T, Yin, P. | | 登録日 | 2017-09-27 | | 公開日 | 2018-06-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.298 Å) | | 主引用文献 | DapF stabilizes the substrate-favoring conformation of RppH to stimulate its RNA-pyrophosphohydrolase activity in Escherichia coli.
Nucleic Acids Res., 46, 2018
|
|
5YZ9
 
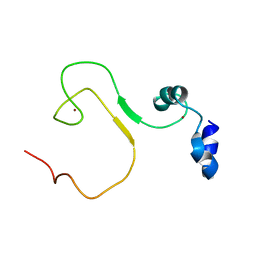 | | zinc finger domain of METTL3-METTL14 N6-methyladenosine methyltransferase | | 分子名称: | N6-adenosine-methyltransferase catalytic subunit, ZINC ION | | 著者 | Dong, X, Tang, C, Gong, Z, Yin, P, Huang, J.B. | | 登録日 | 2017-12-13 | | 公開日 | 2018-03-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR, SOLUTION SCATTERING | | 主引用文献 | Solution structure of the RNA recognition domain of METTL3-METTL14 N6-methyladenosine methyltransferase.
Protein Cell, 10, 2019
|
|
5ZT3
 
 | |
5GP0
 
 | | Crystal structure of geraniol-NUDX1 complex | | 分子名称: | GERANYL DIPHOSPHATE, GLYCEROL, Nudix hydrolase 1 | | 著者 | Liu, J, Guan, Z, Zou, T, Yin, P. | | 登録日 | 2016-07-30 | | 公開日 | 2017-10-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.702 Å) | | 主引用文献 | Structural Insights into the Substrate Recognition Mechanism of Arabidopsis GPP-Bound NUDX1 for Noncanonical Monoterpene Biosynthesis.
Mol Plant, 11, 2018
|
|
5GI0
 
 | | Crystal structure of RNA editing factor MORF9/RIP9 | | 分子名称: | Multiple organellar RNA editing factor 9, chloroplastic | | 著者 | Yan, J, Zhang, Q, Guan, Z, Zou, T, Yin, P. | | 登録日 | 2016-06-21 | | 公開日 | 2017-05-10 | | 実験手法 | X-RAY DIFFRACTION (2.044 Å) | | 主引用文献 | MORF9 increases the RNA-binding activity of PLS-type pentatricopeptide repeat protein in plastid RNA editing
Nat Plants, 3, 2017
|
|
5I9D
 
 | | Crystal structure of designed pentatricopeptide repeat protein dPPR-U8A2 in complex with its target RNA U8A2 | | 分子名称: | RNA (5'-R(*GP*GP*GP*G*UP*UP*UP*UP*AP*AP*UP*UP*UP*UP*CP*CP*CP*C)-3'), pentatricopeptide repeat protein dPPR-U8A2 | | 著者 | Shen, C, Zhang, D, Guan, Z, Zou, T, Yin, P. | | 登録日 | 2016-02-20 | | 公開日 | 2016-04-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.596 Å) | | 主引用文献 | Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins.
Nat Commun, 7, 2016
|
|
5I9H
 
 | | Crystal structure of designed pentatricopeptide repeat protein dPPR-U8G2 in complex with its target RNA U8G2 | | 分子名称: | RNA (5'-R(*GP*GP*GP*GP*UP*UP*UP*UP*GP*GP*UP*UP*UP*UP*CP*CP*CP*C)-3'), pentatricopeptide repeat protein dPPR-U8G2 | | 著者 | Shen, C, Zhang, D, Guan, Z, Zou, T, Yin, P. | | 登録日 | 2016-02-20 | | 公開日 | 2016-04-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.504 Å) | | 主引用文献 | Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins.
Nat Commun, 7, 2016
|
|
5I9G
 
 | | Crystal structure of designed pentatricopeptide repeat protein dPPR-U8C2 in complex with its target RNA U8C2 | | 分子名称: | RNA (5'-R(*GP*GP*G*GP*UP*UP*UP*UP*CP*CP*UP*UP*UP*UP*CP*CP*CP*C)-3'), pentatricopeptide repeat protein dPPR-U8C2 | | 著者 | Shen, C, Zhang, D, Guan, Z, Zou, T, Yin, P. | | 登録日 | 2016-02-20 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.288 Å) | | 主引用文献 | Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins.
Nat Commun, 7, 2016
|
|
6K8K
 
 | | Crystal structure of Arabidopsis thaliana BIC2-CRY2 complex | | 分子名称: | ADENOSINE MONOPHOSPHATE, Cryptochrome-2, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Wang, X, Ma, L, Guan, Z, Yin, P. | | 登録日 | 2019-06-12 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural insights into BIC-mediated inactivation of Arabidopsis cryptochrome 2.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6K8I
 
 | | Crystal structure of Arabidopsis thaliana CRY2 | | 分子名称: | Cryptochrome-2, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Ma, L, Wang, X, Guan, Z, Yin, P. | | 登録日 | 2019-06-12 | | 公開日 | 2020-05-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.697 Å) | | 主引用文献 | Structural insights into BIC-mediated inactivation of Arabidopsis cryptochrome 2.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6KVN
 
 | |
6KVO
 
 | | Crystal structure of chloroplast resolvase in complex with Holliday junction | | 分子名称: | DNA (5'-D(*AP*CP*AP*AP*CP*AP*GP*AP*TP*GP*AP*TP*GP*GP*AP*GP*CP*T)-3'), DNA (5'-D(*GP*CP*CP*TP*TP*GP*CP*TP*TP*GP*GP*AP*CP*AP*TP*CP*TP*T)-3'), DNA (5'-D(P*AP*AP*GP*AP*TP*GP*TP*CP*CP*AP*TP*CP*TP*GP*TP*TP*GP*T)-3'), ... | | 著者 | Yan, J.J, Hong, S.X, Guan, Z.Y, Yin, P. | | 登録日 | 2019-09-05 | | 公開日 | 2020-04-08 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural insights into sequence-dependent Holliday junction resolution by the chloroplast resolvase MOC1.
Nat Commun, 11, 2020
|
|
6LCT
 
 | | Crystal structure of catalytic inactive chloroplast resolvase NtMOC1 in complex with Holliday junction | | 分子名称: | DNA (5'-D(*AP*AP*GP*AP*TP*GP*TP*CP*CP*CP*TP*CP*TP*GP*TP*TP*GP*T)-3'), DNA (5'-D(*AP*CP*AP*AP*CP*AP*GP*AP*GP*GP*AP*TP*GP*GP*AP*GP*CP*T)-3'), DNA (5'-D(*GP*CP*CP*TP*TP*GP*CP*TP*GP*GP*GP*AP*CP*AP*TP*CP*TP*T)-3'), ... | | 著者 | Yan, J.J, Hong, S.X, Guan, Z.Y, Yin, P. | | 登録日 | 2019-11-19 | | 公開日 | 2020-04-08 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structural insights into sequence-dependent Holliday junction resolution by the chloroplast resolvase MOC1.
Nat Commun, 11, 2020
|
|
