4H3S
 
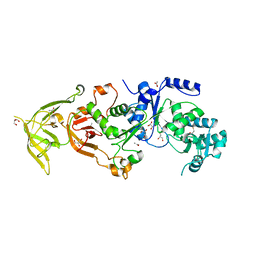 | | The Structure of Glutaminyl-tRNA Synthetase from Saccharomyces Cerevisiae | | 分子名称: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACETATE ION, BROMIDE ION, ... | | 著者 | Snell, E.H, Grant, T.D. | | 登録日 | 2012-09-14 | | 公開日 | 2013-04-24 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | The Structure of Yeast Glutaminyl-tRNA Synthetase and Modeling of Its Interaction with tRNA.
J.Mol.Biol., 425, 2013
|
|
1PZN
 
 | | Rad51 (RadA) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA repair and recombination protein rad51, GLYCEROL, ... | | 著者 | Shin, D.S, Tainer, J.A. | | 登録日 | 2003-07-12 | | 公開日 | 2003-09-09 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Full-length archaeal Rad51 structure and mutants: Mechanisms for RAD51 assembly and control by BRCA2
Embo J., 22, 2003
|
|
4E2F
 
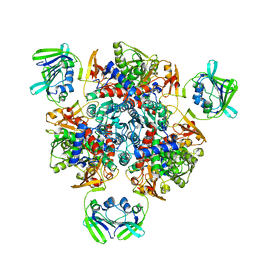 | |
3KFO
 
 | | Crystal structure of the C-terminal domain from the nuclear pore complex component NUP133 from Saccharomyces cerevisiae | | 分子名称: | GLYCEROL, Nucleoporin NUP133 | | 著者 | Sampathkumar, P, Bonanno, J.B, Miller, S, Bain, K, Dickey, M, Gheyi, T, Almo, S.C, Rout, M, Sali, A, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-10-27 | | 公開日 | 2010-01-26 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of the C-terminal domain of Saccharomyces cerevisiae Nup133, a component of the nuclear pore complex.
Proteins, 79, 2011
|
|
3NF5
 
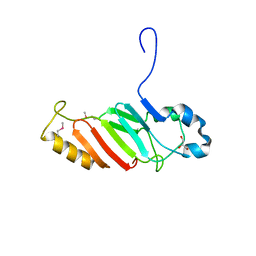 | | Crystal structure of the C-terminal domain of nuclear pore complex component NUP116 from Candida glabrata | | 分子名称: | GLYCEROL, Nucleoporin NUP116 | | 著者 | Sampathkumar, P, Manglicmot, D, Bain, K, Gilmore, J, Gheyi, T, Rout, M, Sali, A, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2010-06-09 | | 公開日 | 2010-08-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Atomic structure of the nuclear pore complex targeting domain of a Nup116 homologue from the yeast, Candida glabrata.
Proteins, 80, 2012
|
|
3KEP
 
 | | Crystal structure of the autoproteolytic domain from the nuclear pore complex component NUP145 from Saccharomyces cerevisiae | | 分子名称: | 1,2-ETHANEDIOL, Nucleoporin NUP145 | | 著者 | Sampathkumar, P, Ozyurt, S.A, Do, J, Bain, K, Dickey, M, Gheyi, T, Sali, A, Kim, S.J, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Rout, M, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-10-26 | | 公開日 | 2009-12-22 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Structures of the autoproteolytic domain from the Saccharomyces cerevisiae nuclear pore complex component, Nup145.
Proteins, 78, 2010
|
|
3KES
 
 | | Crystal structure of the autoproteolytic domain from the nuclear pore complex component NUP145 from Saccharomyces cerevisiae in the Hexagonal, P61 space group | | 分子名称: | 1,2-ETHANEDIOL, Nucleoporin NUP145 | | 著者 | Sampathkumar, P, Ozyurt, S.A, Do, J, Bain, K, Dickey, M, Gheyi, T, Sali, A, Kim, S.J, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Rout, M, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-10-26 | | 公開日 | 2009-12-22 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structures of the autoproteolytic domain from the Saccharomyces cerevisiae nuclear pore complex component, Nup145.
Proteins, 78, 2010
|
|
1OFI
 
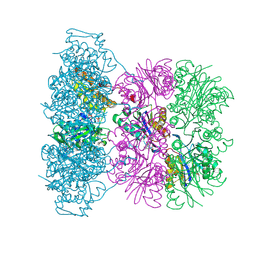 | | Asymmetric complex between HslV and I-domain deleted HslU (H. influenzae) | | 分子名称: | 4-IODO-3-NITROPHENYL ACETYL-LEUCINYL-LEUCINYL-LEUCINYL-VINYLSULFONE, ADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ... | | 著者 | Kwon, A.R, Kessler, B.M, Overkleeft, H.S, McKay, D.B. | | 登録日 | 2003-04-14 | | 公開日 | 2003-07-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structure and Reactivity of an Asymmetric Complex between Hslv and I-Domain Deleted Hslu, a Prokaryotic Homolog of the Eukaryotic Proteasome
J.Mol.Biol., 330, 2003
|
|
1OFH
 
 | | Asymmetric complex between HslV and I-domain deleted HslU (H. influenzae) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV, ... | | 著者 | Kwon, A.R, Kessler, B.M, Overkleeft, H.S, McKay, D.B. | | 登録日 | 2003-04-14 | | 公開日 | 2003-07-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure and Reactivity of an Asymmetric Complex between Hslv and I-Domain Deleted Hslu, a Prokaryotic Homolog of the Eukaryotic Proteasome
J.Mol.Biol., 330, 2003
|
|
1KYI
 
 | | HslUV (H. influenzae)-NLVS Vinyl Sulfone Inhibitor Complex | | 分子名称: | 4-IODO-3-NITROPHENYL ACETYL-LEUCINYL-LEUCINYL-LEUCINYL-VINYLSULFONE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent hsl protease ATP-binding subunit hslU, ... | | 著者 | Sousa, M.C, Kessler, B.M, Overkleeft, H.S, McKay, D.B. | | 登録日 | 2002-02-04 | | 公開日 | 2002-05-15 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Crystal Structure of HslUV Complexed with a Vinyl Sulfone Inhibitor:
Corroboration of a Proposed Mechanism of Allosteric Activation of
HslV by HslU
J.Mol.Biol., 318, 2002
|
|
3BGT
 
 | |
3BH3
 
 | |
3DDX
 
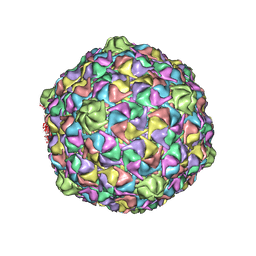 | | HK97 bacteriophage capsid Expansion Intermediate-II model | | 分子名称: | Major capsid protein | | 著者 | Lee, K.K, Gan, L, Conway, J.F, Hendrix, R.W, Steven, A.C, Johnson, J.E. | | 登録日 | 2008-06-06 | | 公開日 | 2008-11-04 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY | | 主引用文献 | Virus capsid expansion driven by the capture of mobile surface loops.
Structure, 16, 2008
|
|
3BH2
 
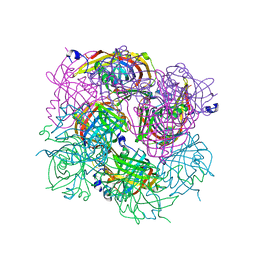 | |
1IN4
 
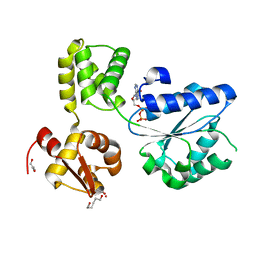 | |
1OHG
 
 | | STRUCTURE OF THE DSDNA BACTERIOPHAGE HK97 MATURE EMPTY CAPSID | | 分子名称: | CHLORIDE ION, MAJOR CAPSID PROTEIN, SULFATE ION | | 著者 | Helgstrand, C, Wikoff, W.R, Duda, R.L, Hendrix, R.W, Johnson, J.E, Liljas, L. | | 登録日 | 2003-05-26 | | 公開日 | 2003-12-11 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (3.45 Å) | | 主引用文献 | The Refined Structure of a Protein Catenane: The Hk97 Bacteriophage Capsid at 3.44A Resolution
J.Mol.Biol., 334, 2003
|
|
1IF0
 
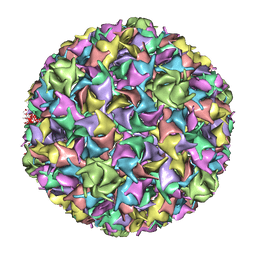 | | PSEUDO-ATOMIC MODEL OF BACTERIOPHAGE HK97 PROCAPSID (PROHEAD II) | | 分子名称: | PROTEIN (MAJOR CAPSID PROTEIN GP5) | | 著者 | Conway, J.F, Wikoff, W.R, Cheng, N, Duda, R.L, Hendrix, R.W, Johnson, J.E, Steven, A.C. | | 登録日 | 2001-04-11 | | 公開日 | 2001-05-02 | | 最終更新日 | 2024-02-07 | | 実験手法 | ELECTRON MICROSCOPY (12 Å) | | 主引用文献 | Virus maturation involving large subunit rotations and local refolding.
Science, 292, 2001
|
|
1IM2
 
 | | HslU, Haemophilus Influenzae, Selenomethionine Variant | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, SULFATE ION | | 著者 | Trame, C.B, McKay, D.B. | | 登録日 | 2001-05-09 | | 公開日 | 2001-08-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of Haemophilus influenzae HslU protein in crystals with one-dimensional disorder twinning.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
