3HRU
 
 | | Crystal Structure of ScaR with bound Zn2+ | | 分子名称: | Metalloregulator ScaR, SULFATE ION, ZINC ION | | 著者 | Stoll, K.E, Draper, W.E, Kliegman, J.I, Golynskiy, M.V, Brew-Appiah, R.A.T, Brown, H.K, Breyer, W.A, Jakubovics, N.S, Jenkinson, H.F, Brennan, R.B, Cohen, S.M, Glasfeld, A. | | 登録日 | 2009-06-09 | | 公開日 | 2009-06-23 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Characterization and structure of the manganese-responsive transcriptional regulator ScaR.
Biochemistry, 48, 2009
|
|
3HRT
 
 | | Crystal Structure of ScaR with bound Cd2+ | | 分子名称: | CADMIUM ION, Metalloregulator ScaR, SULFATE ION | | 著者 | Stoll, K.E, Draper, W.E, Kliegman, J.I, Golynskiy, M.V, Brew-Appiah, R.A.T, Brown, H.K, Breyer, W.A, Jakubovics, N.S, Jenkinson, H.F, Brennan, R.B, Cohen, S.M, Glasfeld, A. | | 登録日 | 2009-06-09 | | 公開日 | 2009-06-23 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Characterization and structure of the manganese-responsive transcriptional regulator ScaR.
Biochemistry, 48, 2009
|
|
3HRS
 
 | | Crystal Structure of the Manganese-activated Repressor ScaR: apo form | | 分子名称: | Metalloregulator ScaR, SULFATE ION | | 著者 | Stoll, K.E, Draper, W.E, Kliegman, J.I, Golynskiy, M.V, Brew-Appiah, R.A.T, Brown, H.K, Breyer, W.A, Jakubovics, N.S, Jenkinson, H.F, Brennan, R.B, Cohen, S.M, Glasfeld, A. | | 登録日 | 2009-06-09 | | 公開日 | 2009-06-23 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Characterization and structure of the manganese-responsive transcriptional regulator ScaR.
Biochemistry, 48, 2009
|
|
6K8W
 
 | |
6K8S
 
 | |
6K8U
 
 | |
6K8V
 
 | |
6IJ6
 
 | |
6IJ4
 
 | |
6IJ3
 
 | |
6IJ5
 
 | |
6K8T
 
 | |
5ZRD
 
 | | Tyrosinase from Burkholderia thailandensis (BtTYR) at low pH condition | | 分子名称: | CITRIC ACID, COPPER (II) ION, GLYCEROL, ... | | 著者 | Lee, S, Son, H.-F, Kim, K.-J. | | 登録日 | 2018-04-24 | | 公開日 | 2018-10-31 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Basis for Highly Efficient Production of Catechol Derivatives at Acidic pH by Tyrosinase from Burkholderia thailandensis
Acs Catalysis, 8, 2018
|
|
5ZRE
 
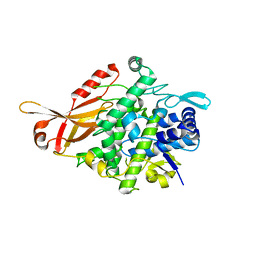 | | Tyrosinase from Burkholderia thailandensis (BtTYR) at high pH condition | | 分子名称: | COPPER (II) ION, GLYCEROL, OXYGEN ATOM, ... | | 著者 | Lee, S, Son, H.-F, Kim, K.-J. | | 登録日 | 2018-04-24 | | 公開日 | 2018-10-31 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural Basis for Highly Efficient Production of Catechol Derivatives at Acidic pH by Tyrosinase from Burkholderia thailandensis
Acs Catalysis, 8, 2018
|
|
5X7N
 
 | |
5X7M
 
 | |
5X5T
 
 | |
5X5U
 
 | |
5DZA
 
 | | Streptococcus agalactiae AgI/II polypeptide BspA C terminal domain (WT) | | 分子名称: | 1,2-ETHANEDIOL, BspA, DI(HYDROXYETHYL)ETHER | | 著者 | Rego, S, Till, M, Race, P.R. | | 登録日 | 2015-09-25 | | 公開日 | 2016-06-22 | | 最終更新日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Structural and Functional Analysis of Cell Wall-anchored Polypeptide Adhesin BspA in Streptococcus agalactiae.
J.Biol.Chem., 291, 2016
|
|
6ABY
 
 | | Crystal structure of citrate synthase (Msed_1522) from Metallosphaera sedula in complex with oxaloacetate | | 分子名称: | ACETYL COENZYME *A, Citrate synthase, GLYCEROL, ... | | 著者 | Lee, S.-H, Son, H.-F, Kim, K.-J. | | 登録日 | 2018-07-24 | | 公開日 | 2019-03-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural insights into the inhibition properties of archaeon citrate synthase from Metallosphaera sedula.
PLoS ONE, 14, 2019
|
|
6ABX
 
 | |
4NZS
 
 | |
5GJN
 
 | |
5GJM
 
 | |
5GJP
 
 | |
