1KFA
 
 | | Crystal structure of Fab fragment complexed with gibberellin A4 | | 分子名称: | GIBBERELLIN A4, monoclonal antibody heavy chain, monoclonal antibody light chain | | 著者 | Murata, T, Fushinobu, S, Nakajima, M, Asami, O, Sassa, T, Wakagi, T, Yamaguchi, I. | | 登録日 | 2001-11-20 | | 公開日 | 2002-09-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of the liganded anti-gibberellin A(4) antibody 4-B8(8)/E9 Fab fragment.
Biochem.Biophys.Res.Commun., 293, 2002
|
|
5GZH
 
 | | Endo-beta-1,2-glucanase from Chitinophaga pinensis - ligand free form | | 分子名称: | Endo-beta-1,2-glucanase, IODIDE ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | 著者 | Abe, K, Nakajima, M, Arakawa, T, Fushinobu, S, Taguchi, H. | | 登録日 | 2016-09-28 | | 公開日 | 2017-03-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Biochemical and structural analyses of a bacterial endo-beta-1,2-glucanase reveal a new glycoside hydrolase family
J. Biol. Chem., 292, 2017
|
|
5GZK
 
 | | Endo-beta-1,2-glucanase from Chitinophaga pinensis - sophorotriose and glucose complex | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, ... | | 著者 | Abe, K, Nakajima, M, Arakawa, T, Fushinobu, S, Taguchi, H. | | 登録日 | 2016-09-28 | | 公開日 | 2017-03-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Biochemical and structural analyses of a bacterial endo-beta-1,2-glucanase reveal a new glycoside hydrolase family
J. Biol. Chem., 292, 2017
|
|
5YSD
 
 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in complex with sophorotriose | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lin1841 protein, MAGNESIUM ION, ... | | 著者 | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | 登録日 | 2017-11-14 | | 公開日 | 2018-05-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
5YSF
 
 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in complex with sophoropentaose | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lin1841 protein, MAGNESIUM ION, ... | | 著者 | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | 登録日 | 2017-11-14 | | 公開日 | 2018-05-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
5YSE
 
 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in complex with sophorotetraose | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lin1841 protein, MAGNESIUM ION, ... | | 著者 | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | 登録日 | 2017-11-14 | | 公開日 | 2018-05-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
5YSB
 
 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in ligand-free form | | 分子名称: | DI(HYDROXYETHYL)ETHER, Lin1841 protein, ZINC ION | | 著者 | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | 登録日 | 2017-11-13 | | 公開日 | 2018-05-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
3WFJ
 
 | | The complex structure of D-mandelate dehydrogenase with NADH | | 分子名称: | 2-dehydropantoate 2-reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Miyanaga, A, Fujisawa, S, Furukawa, N, Arai, K, Nakajima, M, Taguchi, H. | | 登録日 | 2013-07-19 | | 公開日 | 2014-07-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The crystal structure of D-mandelate dehydrogenase reveals its distinct substrate and coenzyme recognition mechanisms from those of 2-ketopantoate reductase.
Biochem.Biophys.Res.Commun., 439, 2013
|
|
3WFI
 
 | | The crystal structure of D-mandelate dehydrogenase | | 分子名称: | 2-dehydropantoate 2-reductase | | 著者 | Miyanaga, A, Fujisawa, S, Furukawa, N, Arai, K, Nakajima, M, Taguchi, H. | | 登録日 | 2013-07-19 | | 公開日 | 2014-07-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.997 Å) | | 主引用文献 | The crystal structure of D-mandelate dehydrogenase reveals its distinct substrate and coenzyme recognition mechanisms from those of 2-ketopantoate reductase.
Biochem.Biophys.Res.Commun., 439, 2013
|
|
7WCQ
 
 | | Crystal structure of HIV-1 protease in complex with lactam derivative 1 | | 分子名称: | (3R,4R)-3-[(4-fluorophenyl)methyl]-1-[(4-methoxyphenyl)methyl]-3-(4-methylsulfonylphenyl)-4-oxidanyl-pyrrolidin-2-one, Protease | | 著者 | Kojima, E, Iimuro, A, Nakajima, M, Kinuta, H, Asada, N, Sako, Y, Nakata, Z, Uemura, K, Arita, S, Miki, S, Wakasa-Morimoto, C, Tachibana, Y, Fumoto, M. | | 登録日 | 2021-12-20 | | 公開日 | 2022-11-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.011 Å) | | 主引用文献 | Pocket-to-Lead: Structure-Based De Novo Design of Novel Non-peptidic HIV-1 Protease Inhibitors Using the Ligand Binding Pocket as a Template.
J.Med.Chem., 65, 2022
|
|
7WBS
 
 | | Crystal structure of HIV-1 protease in complex with lactam derivative 2 | | 分子名称: | (3~{R},4~{R})-1-[(4-methoxyphenyl)methyl]-3-(3-methylbutyl)-3-[4-methylsulfonyl-2-[(2~{S})-1-oxidanylpropan-2-yl]oxy-phenyl]-4-oxidanyl-pyrrolidin-2-one, GLYCEROL, Protease | | 著者 | Kojima, E, Iimuro, A, Nakajima, M, Kinuta, H, Asada, N, Sako, Y, Nakata, Z, Uemura, K, Arita, S, Miki, S, Wakabayashi-Morimoto, C, Tachibana, Y, Fumoto, M. | | 登録日 | 2021-12-17 | | 公開日 | 2022-11-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Pocket-to-Lead: Structure-Based De Novo Design of Novel Non-peptidic HIV-1 Protease Inhibitors Using the Ligand Binding Pocket as a Template.
J.Med.Chem., 65, 2022
|
|
5X20
 
 | | The ternary structure of D-mandelate dehydrogenase with NADH and anilino(oxo)acetate | | 分子名称: | 2-dehydropantoate 2-reductase, 2-oxidanylidene-2-phenylazanyl-ethanoic acid, GLYCEROL, ... | | 著者 | Furukawa, N, Miyanaga, A, Nakajima, M, Taguchi, H. | | 登録日 | 2017-01-29 | | 公開日 | 2017-04-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The ternary complex structure of d-mandelate dehydrogenase with NADH and anilino(oxo)acetate.
Biochem. Biophys. Res. Commun., 486, 2017
|
|
5Z20
 
 | | The ternary structure of D-lactate dehydrogenase from Pseudomonas aeruginosa with NADH and oxamate | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, D-lactate dehydrogenase (Fermentative), DI(HYDROXYETHYL)ETHER, ... | | 著者 | Furukawa, N, Miyanaga, A, Nakajima, M, Taguchi, H. | | 登録日 | 2017-12-28 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural Basis of Sequential Allosteric Transitions in Tetrameric d-Lactate Dehydrogenases from Three Gram-Negative Bacteria.
Biochemistry, 57, 2018
|
|
5Z06
 
 | | Crystal structure of beta-1,2-glucanase from Parabacteroides distasonis | | 分子名称: | BDI_3064 protein, CALCIUM ION, GLYCEROL | | 著者 | Shimizu, H, Nakajima, M, Miyanaga, A, Takahashi, Y, Tanaka, N, Kobayashi, K, Sugimoto, N, Nakai, H, Taguchi, H. | | 登録日 | 2017-12-18 | | 公開日 | 2018-05-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Characterization and Structural Analysis of a Novel exo-Type Enzyme Acting on beta-1,2-Glucooligosaccharides from Parabacteroides distasonis
Biochemistry, 57, 2018
|
|
5Z21
 
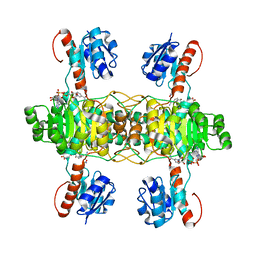 | | The ternary structure of D-lactate dehydrogenase from Fusobacterium nucleatum with NADH and oxamate | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, D-lactate dehydrogenase, OXAMIC ACID | | 著者 | Furukawa, N, Miyanaga, A, Nakajima, M, Taguchi, H. | | 登録日 | 2017-12-28 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Basis of Sequential Allosteric Transitions in Tetrameric d-Lactate Dehydrogenases from Three Gram-Negative Bacteria.
Biochemistry, 57, 2018
|
|
5Z1Z
 
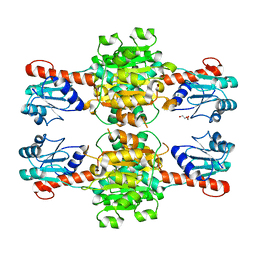 | | The apo-structure of D-lactate dehydrogenase from Escherichia coli | | 分子名称: | D-isomer specific 2-hydroxyacid dehydrogenase NAD-binding, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION | | 著者 | Furukawa, N, Miyanaga, A, Nakajima, M, Taguchi, H. | | 登録日 | 2017-12-28 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Structural Basis of Sequential Allosteric Transitions in Tetrameric d-Lactate Dehydrogenases from Three Gram-Negative Bacteria.
Biochemistry, 57, 2018
|
|
6ABI
 
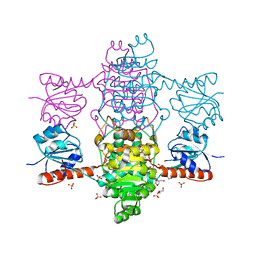 | | The apo-structure of D-lactate dehydrogenase from Fusobacterium nucleatum | | 分子名称: | D-lactate dehydrogenase, GLYCEROL, SULFATE ION | | 著者 | Furukawa, N, Miyanaga, A, Nakajima, M, Taguchi, H. | | 登録日 | 2018-07-21 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Basis of Sequential Allosteric Transitions in Tetrameric d-Lactate Dehydrogenases from Three Gram-Negative Bacteria
Biochemistry, 57, 2018
|
|
6ABJ
 
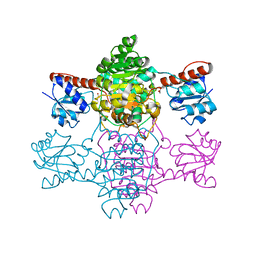 | | The apo-structure of D-lactate dehydrogenase from Pseudomonas aeruginosa | | 分子名称: | ACETATE ION, D-lactate dehydrogenase (Fermentative) | | 著者 | Furukawa, N, Miyanaga, A, Nakajima, M, Taguchi, H. | | 登録日 | 2018-07-21 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Structural Basis of Sequential Allosteric Transitions in Tetrameric d-Lactate Dehydrogenases from Three Gram-Negative Bacteria
Biochemistry, 57, 2018
|
|
7VL0
 
 | | The complex structure of beta-1,2-glucosyltransferase from Ignavibacterium album with p-nitrophenyl-alpha-D-glucopyranoside | | 分子名称: | 4-nitrophenyl alpha-D-glucopyranoside, Beta-galactosidase, CALCIUM ION | | 著者 | Kobayashi, K, Shimizu, H, Tanaka, N, Kuramochi, K, Nakai, H, Nakajima, M, Taguchi, H. | | 登録日 | 2021-10-01 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Characterization and structural analyses of a novel glycosyltransferase acting on the beta-1,2-glucosidic linkages.
J.Biol.Chem., 298, 2022
|
|
7VL3
 
 | | The complex structure of beta-1,2-glucosyltransferase from Ignavibacterium album with phenyl alpha-D-glucoside | | 分子名称: | (2R,3S,4S,5R,6R)-2-(hydroxymethyl)-6-phenoxy-oxane-3,4,5-triol, CALCIUM ION, beta-1,2-glucosyltransferase | | 著者 | Kobayashi, K, Shimizu, H, Tanaka, N, Kuramochi, K, Nakai, H, Nakajima, M, Taguchi, H. | | 登録日 | 2021-10-01 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Characterization and structural analyses of a novel glycosyltransferase acting on the beta-1,2-glucosidic linkages.
J.Biol.Chem., 298, 2022
|
|
7VL7
 
 | | The complex structure of beta-1,2-glucosyltransferase from Ignavibacterium album with esculin | | 分子名称: | 6-[(2S,3R,4S,5S,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-7-oxidanyl-chromen-2-one, CALCIUM ION, beta-1,2-glucosyltransferase | | 著者 | Kobayashi, K, Shimizu, H, Tanaka, N, Kuramochi, K, Nakai, H, Nakajima, M, Taguchi, H. | | 登録日 | 2021-10-01 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Characterization and structural analyses of a novel glycosyltransferase acting on the beta-1,2-glucosidic linkages.
J.Biol.Chem., 298, 2022
|
|
7VKY
 
 | | The complex structure of beta-1,2-glucosyltransferase from Ignavibacterium album with sophorose | | 分子名称: | CALCIUM ION, beta-1,2-glucosyltransferase, beta-D-glucopyranose-(1-2)-alpha-D-glucopyranose, ... | | 著者 | Kobayashi, K, Shimizu, H, Tanaka, N, Kuramochi, K, Nakai, H, Nakajima, M, Taguchi, H. | | 登録日 | 2021-10-01 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Characterization and structural analyses of a novel glycosyltransferase acting on the beta-1,2-glucosidic linkages.
J.Biol.Chem., 298, 2022
|
|
7VL5
 
 | | The complex structure of beta-1,2-glucosyltransferase from Ignavibacterium album with n-octyl-beta-D-glucoside | | 分子名称: | Beta-galactosidase, CALCIUM ION, octyl beta-D-glucopyranoside | | 著者 | Kobayashi, K, Shimizu, H, Tanaka, N, Kuramochi, K, Nakai, H, Nakajima, M, Taguchi, H. | | 登録日 | 2021-10-01 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Characterization and structural analyses of a novel glycosyltransferase acting on the beta-1,2-glucosidic linkages.
J.Biol.Chem., 298, 2022
|
|
7VL4
 
 | | The complex structure of beta-1,2-glucosyltransferase from Ignavibacterium album with methyl beta-D-glucoside | | 分子名称: | CALCIUM ION, beta-1,2-glucosyltransferase, methyl beta-D-glucopyranoside | | 著者 | Kobayashi, K, Shimizu, H, Tanaka, N, Kuramochi, K, Nakai, H, Nakajima, M, Taguchi, H. | | 登録日 | 2021-10-01 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Characterization and structural analyses of a novel glycosyltransferase acting on the beta-1,2-glucosidic linkages.
J.Biol.Chem., 298, 2022
|
|
7VL2
 
 | | The complex structure of beta-1,2-glucosyltransferase from Ignavibacterium album with ethyl alpha-D-Glucoside | | 分子名称: | (2~{S},3~{R},4~{S},5~{S},6~{R})-2-ethoxy-6-(hydroxymethyl)oxane-3,4,5-triol, CALCIUM ION, beta-1,2-glucosyltransferase | | 著者 | Kobayashi, K, Shimizu, H, Tanaka, N, Kuramochi, K, Nakai, H, Nakajima, M, Taguchi, H. | | 登録日 | 2021-10-01 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Characterization and structural analyses of a novel glycosyltransferase acting on the beta-1,2-glucosidic linkages.
J.Biol.Chem., 298, 2022
|
|
