3CNL
 
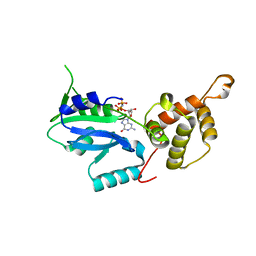 | | Crystal structure of GNP-bound YlqF from T. maritima | | 分子名称: | PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Putative uncharacterized protein | | 著者 | Kim, D.J, Jang, J.Y, Yoon, H.-J, Suh, S.W. | | 登録日 | 2008-03-26 | | 公開日 | 2008-06-24 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of YlqF, a circularly permuted GTPase: Implications for its GTPase activation in 50 S ribosomal subunit assembly
Proteins, 72, 2008
|
|
3OBY
 
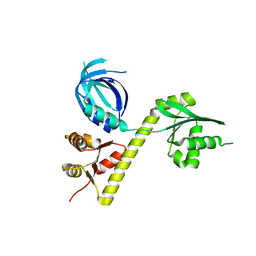 | | Crystal structure of Archaeoglobus fulgidus Pelota reveals inter-domain structural plasticity | | 分子名称: | Protein pelota homolog | | 著者 | Lee, H.H, Jang, J.Y, Yoon, H.-J, Kim, S.J, Suh, S.W. | | 登録日 | 2010-08-09 | | 公開日 | 2010-09-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structures of two archaeal Pelotas reveal inter-domain structural plasticity
Biochem.Biophys.Res.Commun., 399, 2010
|
|
3OBW
 
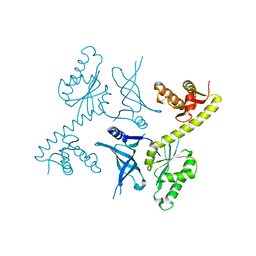 | | Crystal structure of two archaeal Pelotas reveal inter-domain structural plasticity | | 分子名称: | Protein pelota homolog | | 著者 | Lee, H.H, Jang, J.Y, Yoon, H.-J, Kim, S.J, Suh, S.W. | | 登録日 | 2010-08-09 | | 公開日 | 2010-09-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structures of two archaeal Pelotas reveal inter-domain structural plasticity
Biochem.Biophys.Res.Commun., 399, 2010
|
|
8I28
 
 | |
1KBT
 
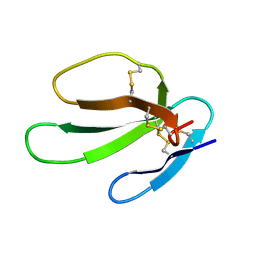 | | SOLUTION STRUCTURE OF CARDIOTOXIN IV, NMR, 12 STRUCTURES | | 分子名称: | CTX IV | | 著者 | Jeng, J.Y, Kumar, T.K.S, Jayaraman, G, Yu, C. | | 登録日 | 1996-07-22 | | 公開日 | 1997-07-23 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comparison of the hemolytic activity and solution structures of two snake venom cardiotoxin analogues which only differ in their N-terminal amino acid.
Biochemistry, 36, 1997
|
|
1KBS
 
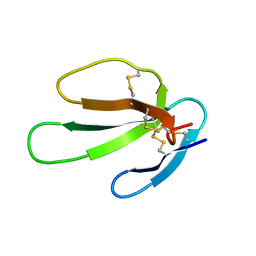 | | SOLUTION STRUCTURE OF CARDIOTOXIN IV, NMR, 1 STRUCTURE | | 分子名称: | CTX IV | | 著者 | Jeng, J.Y, Kumar, T.K.S, Jayaraman, G, Yu, C. | | 登録日 | 1996-07-22 | | 公開日 | 1997-07-23 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comparison of the hemolytic activity and solution structures of two snake venom cardiotoxin analogues which only differ in their N-terminal amino acid.
Biochemistry, 36, 1997
|
|
4XZZ
 
 | | Structure of Helicobacter pylori Csd6 in the ligand-free state | | 分子名称: | Conserved hypothetical secreted protein, GLYCEROL | | 著者 | Kim, H.S, Im, H.N, Yoon, H.J, Suh, S.W. | | 登録日 | 2015-02-05 | | 公開日 | 2015-09-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | The Cell Shape-determining Csd6 Protein from Helicobacter pylori Constitutes a New Family of l,d-Carboxypeptidase
J.Biol.Chem., 290, 2015
|
|
4Y4V
 
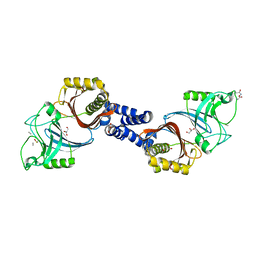 | | Structure of Helicobacter pylori Csd6 in the D-Ala-bound state | | 分子名称: | Conserved hypothetical secreted protein, D-ALANINE, GLYCEROL | | 著者 | Kim, H.S, Im, H.N, Yoon, H.J, Suh, S.W. | | 登録日 | 2015-02-11 | | 公開日 | 2015-09-02 | | 最終更新日 | 2022-03-23 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | The Cell Shape-determining Csd6 Protein from Helicobacter pylori Constitutes a New Family of l,d-Carboxypeptidase
J.Biol.Chem., 290, 2015
|
|
3AKK
 
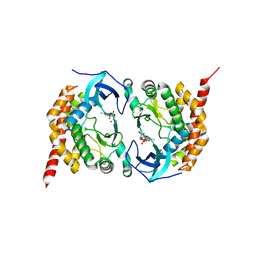 | |
3AKJ
 
 | |
3AKL
 
 | |
6L4H
 
 | |
6L4B
 
 | | Crystal structure of human WT NDRG3 | | 分子名称: | Protein NDRG3 | | 著者 | Kim, K.R, Han, B.W. | | 登録日 | 2019-10-16 | | 公開日 | 2020-08-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural and Biophysical Analyses of Human N-Myc Downstream-Regulated Gene 3 (NDRG3) Protein.
Biomolecules, 10, 2020
|
|
5J1M
 
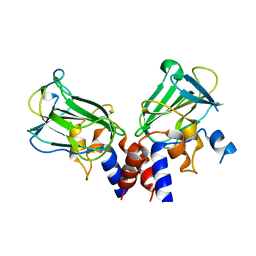 | | Crystal structure of Csd1-Csd2 dimer II | | 分子名称: | ToxR-activated gene (TagE), ZINC ION | | 著者 | An, D.R, Suh, S.W. | | 登録日 | 2016-03-29 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural Basis of the Heterodimer Formation between Cell Shape-Determining Proteins Csd1 and Csd2 from Helicobacter pylori
Plos One, 11, 2016
|
|
5J1K
 
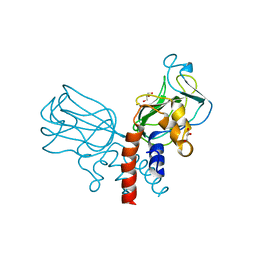 | | Crystal structure of Csd2-Csd2 dimer | | 分子名称: | GLYCEROL, ToxR-activated gene (TagE) | | 著者 | An, D.R, Suh, S.W. | | 登録日 | 2016-03-29 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Structural Basis of the Heterodimer Formation between Cell Shape-Determining Proteins Csd1 and Csd2 from Helicobacter pylori
Plos One, 11, 2016
|
|
5J1L
 
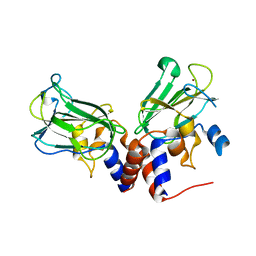 | | Crystal structure of Csd1-Csd2 dimer I | | 分子名称: | ToxR-activated gene (TagE), ZINC ION | | 著者 | An, D.R, Suh, S.W. | | 登録日 | 2016-03-29 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Structural Basis of the Heterodimer Formation between Cell Shape-Determining Proteins Csd1 and Csd2 from Helicobacter pylori
Plos One, 11, 2016
|
|
8JJW
 
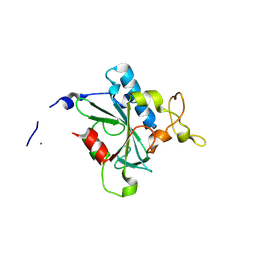 | | Crystal structure of QG-hNTAQ1 C28S | | 分子名称: | MAGNESIUM ION, Protein N-terminal glutamine amidohydrolase | | 著者 | Kang, J.M, Han, B.W. | | 登録日 | 2023-05-31 | | 公開日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JJY
 
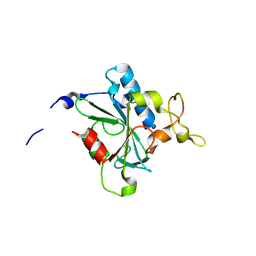 | | Crystal structure of QN-hNTAQ1 C28S | | 分子名称: | Protein N-terminal glutamine amidohydrolase | | 著者 | Kang, J.M, Han, B.W. | | 登録日 | 2023-05-31 | | 公開日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JK2
 
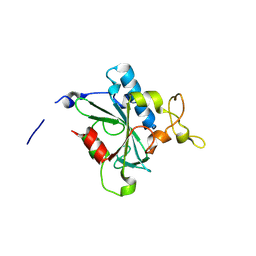 | | Crystal structure of QF-hNTAQ1 C28S | | 分子名称: | Protein N-terminal glutamine amidohydrolase | | 著者 | Kang, J.M, Han, B.W. | | 登録日 | 2023-05-31 | | 公開日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.742 Å) | | 主引用文献 | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JJG
 
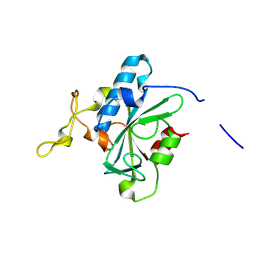 | | Crystal structure of QW-hNTAQ1 C28S | | 分子名称: | Protein N-terminal glutamine amidohydrolase | | 著者 | Kang, J.M, Han, B.W. | | 登録日 | 2023-05-30 | | 公開日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JJI
 
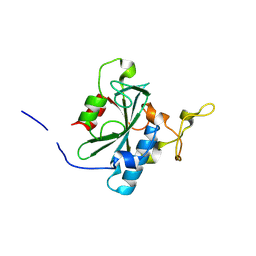 | | Crystal structure of QR-hNTAQ1 C28S | | 分子名称: | Protein N-terminal glutamine amidohydrolase | | 著者 | Kang, J.M, Han, B.W. | | 登録日 | 2023-05-30 | | 公開日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (2.206 Å) | | 主引用文献 | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JK0
 
 | | Crystal structure of QL-hNTAQ1 C28S | | 分子名称: | Protein N-terminal glutamine amidohydrolase | | 著者 | Kang, J.M, Han, B.W. | | 登録日 | 2023-05-31 | | 公開日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JJH
 
 | | Crystal structure of QH-hNTAQ1 C28S | | 分子名称: | Protein N-terminal glutamine amidohydrolase | | 著者 | Kang, J.M, Han, B.W. | | 登録日 | 2023-05-30 | | 公開日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JJX
 
 | | Crystal structure of QS-hNTAQ1 C28S | | 分子名称: | Protein N-terminal glutamine amidohydrolase | | 著者 | Kang, J.M, Han, B.W. | | 登録日 | 2023-05-31 | | 公開日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JJF
 
 | | Crystal structure of QE-hNTAQ1 C28S | | 分子名称: | Protein N-terminal glutamine amidohydrolase | | 著者 | Kang, J.M, Han, B.W. | | 登録日 | 2023-05-30 | | 公開日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
