1RDB
 
 | |
1RDC
 
 | |
1RDA
 
 | |
1KVA
 
 | | E. COLI RIBONUCLEASE HI D134A MUTANT | | 分子名称: | RIBONUCLEASE H | | 著者 | Kashiwagi, T, Jeanteur, D, Haruki, M, Katayanagi, K, Kanaya, S, Morikawa, K. | | 登録日 | 1996-10-04 | | 公開日 | 1997-03-12 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Proposal for new catalytic roles for two invariant residues in Escherichia coli ribonuclease HI.
Protein Eng., 9, 1996
|
|
1KVC
 
 | | E. COLI RIBONUCLEASE HI D134N MUTANT | | 分子名称: | RIBONUCLEASE H | | 著者 | Kashiwagi, T, Jeanteur, D, Haruki, M, Katayanagi, K, Kanaya, S, Morikawa, K. | | 登録日 | 1996-10-04 | | 公開日 | 1997-03-12 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Proposal for new catalytic roles for two invariant residues in Escherichia coli ribonuclease HI.
Protein Eng., 9, 1996
|
|
1KVB
 
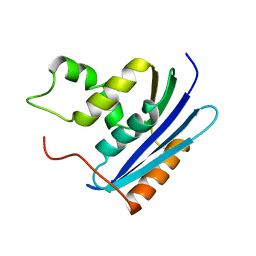 | | E. COLI RIBONUCLEASE HI D134H MUTANT | | 分子名称: | RIBONUCLEASE H | | 著者 | Kashiwagi, T, Jeanteur, D, Haruki, M, Katayanagi, K, Kanaya, S, Morikawa, K. | | 登録日 | 1996-10-04 | | 公開日 | 1997-03-12 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Proposal for new catalytic roles for two invariant residues in Escherichia coli ribonuclease HI.
Protein Eng., 9, 1996
|
|
1RDD
 
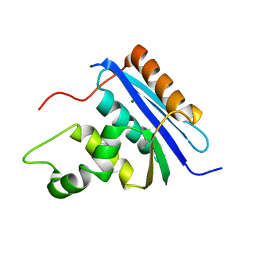 | |
3WRY
 
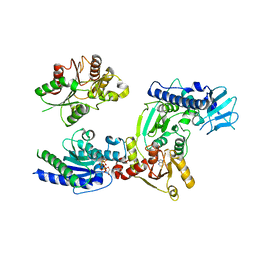 | | Crystal structure of helicase complex 2 | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Matsumura, H, Katoh, E. | | 登録日 | 2014-02-27 | | 公開日 | 2014-08-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis for the recognition-evasion arms race between Tomato mosaic virus and the resistance gene Tm-1
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WRX
 
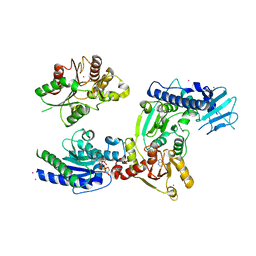 | | Crystal structure of helicase complex 1 | | 分子名称: | CESIUM ION, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Matsumura, H, Katoh, E. | | 登録日 | 2014-02-27 | | 公開日 | 2014-08-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis for the recognition-evasion arms race between Tomato mosaic virus and the resistance gene Tm-1
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WRV
 
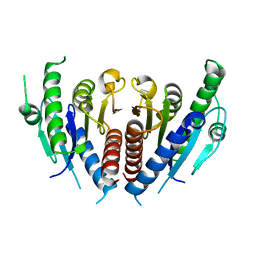 | |
5GVI
 
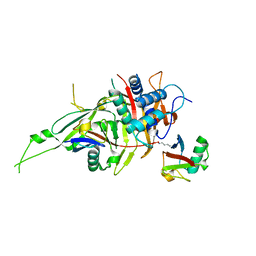 | |
1BJ3
 
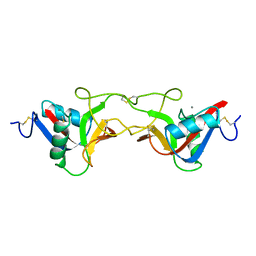 | | CRYSTAL STRUCTURE OF COAGULATION FACTOR IX-BINDING PROTEIN (IX-BP) FROM VENOM OF HABU SNAKE WITH A HETERODIMER OF C-TYPE LECTIN DOMAINS | | 分子名称: | CALCIUM ION, PROTEIN (COAGULATION FACTOR IX-BINDING PROTEIN A), PROTEIN (COAGULATION FACTOR IX-BINDING PROTEIN B) | | 著者 | Mizuno, H, Fujimoto, Z, Koizumi, M, Kano, H, Atoda, H, Morita, T. | | 登録日 | 1998-07-02 | | 公開日 | 1999-08-16 | | 最終更新日 | 2017-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of coagulation factor IX-binding protein from habu snake venom at 2.6 A: implication of central loop swapping based on deletion in the linker region.
J.Mol.Biol., 289, 1999
|
|
7E9S
 
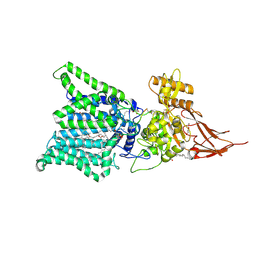 | | Archaeal oligosaccharyltransferase AglB from Archaeoglobus fulgidus in complex with an inhibitory peptide and a dolichol-phosphate | | 分子名称: | (2R)-2,3-DIHYDROXYPROPYL (7Z)-TETRADEC-7-ENOATE, DI(HYDROXYETHYL)ETHER, Dolichyl-phosphooligosaccharide-protein glycotransferase 3, ... | | 著者 | Taguchi, Y, Hirata, K, Kohda, D. | | 登録日 | 2021-03-05 | | 公開日 | 2021-09-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The structure of an archaeal oligosaccharyltransferase provides insight into the strict exclusion of proline from the N-glycosylation sequon.
Commun Biol, 4, 2021
|
|
6K7F
 
 | |
6K7E
 
 | |
6K8Q
 
 | | Solution structure of the intermembrane space domain of the mitochondrial import protein Tim21 from S. cerevisiae | | 分子名称: | Mitochondrial import inner membrane translocase subunit TIM21 | | 著者 | Bala, S, Shinya, S, Srivastava, A, Shimada, A, Kobayashi, N, Kojima, C, Tama, F, Miyashita, O, Kohda, D. | | 登録日 | 2019-06-13 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Crystal contact-free conformation of an intrinsically flexible loop in protein crystal: Tim21 as the case study.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
6K7D
 
 | |
6JQA
 
 | |
7WBK
 
 | |
7WBM
 
 | | Crystal structure of Legionella pneumophila effector protein Lpg0081 | | 分子名称: | Lpg0081, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | 著者 | Lee, J, Kim, H, Oh, B.H. | | 登録日 | 2021-12-17 | | 公開日 | 2022-06-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Reversible modification of mitochondrial ADP/ATP translocases by paired Legionella effector proteins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
