8H33
 
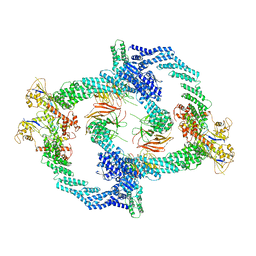 | | Cryo-EM Structure of the KBTBD2-Cul3-Rbx1 tetrameric complex | | 分子名称: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | 著者 | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | 登録日 | 2022-10-07 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (7.86 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H34
 
 | | Cryo-EM Structure of the KBTBD2-Cul3-Rbx1 hexameric complex | | 分子名称: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | 著者 | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | 登録日 | 2022-10-07 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (7.99 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H37
 
 | | Cryo-EM Structure of the KBTBD2-CUL3-Rbx1-p85a tetrameric complex | | 分子名称: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | 著者 | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | 登録日 | 2022-10-08 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (7.52 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H3R
 
 | | Cryo-EM Structure of the KBTBD2-CRL3~N8 dimeric complex | | 分子名称: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | 著者 | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | 登録日 | 2022-10-09 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (6.36 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H38
 
 | | Cryo-EM Structure of the KBTBD2-CRL3~N8-CSN(mutate) complex | | 分子名称: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | 著者 | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | 登録日 | 2022-10-08 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.25 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
9C6M
 
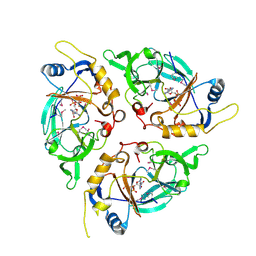 | | Yasminevirus c12orf29, a 5' to 3' RNA ligase, K73M mutant | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, RNA ligase1 | | 著者 | Hu, Y, Lopez, V.A, Tagliabracci, V.S, Tomchick, D.R. | | 登録日 | 2024-06-07 | | 公開日 | 2024-10-02 | | 最終更新日 | 2025-03-05 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Biochemical and structural insights into a 5' to 3' RNA ligase reveal a potential role in tRNA ligation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9C6L
 
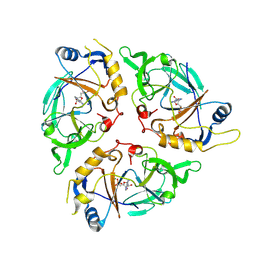 | | Yasminevirus c12orf29, a 5' to 3' RNA ligase | | 分子名称: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, RNA ligase1, ... | | 著者 | Hu, Y, Lopez, V.A, Tagliabracci, V.S, Tomchick, D.R. | | 登録日 | 2024-06-07 | | 公開日 | 2024-10-02 | | 最終更新日 | 2025-03-05 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Biochemical and structural insights into a 5' to 3' RNA ligase reveal a potential role in tRNA ligation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9DEU
 
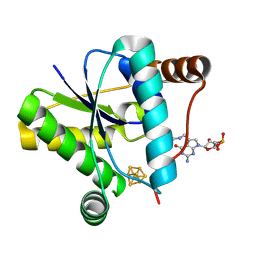 | | Crystal structure of epoxyqueuosine reductase QueH in complex with queuosine | | 分子名称: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-7-(5-O-phosphono-beta-D-ribofuranosyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, CHLORIDE ION, Epoxyqueuosine reductase QueH, ... | | 著者 | Hu, Y, Bruner, S.D. | | 登録日 | 2024-08-29 | | 公開日 | 2024-12-18 | | 最終更新日 | 2025-02-05 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Mechanism of Catalysis and Substrate Binding of Epoxyqueuosine Reductase in the Biosynthetic Pathway to Queuosine-Modified tRNA.
Biochemistry, 64, 2025
|
|
9EZZ
 
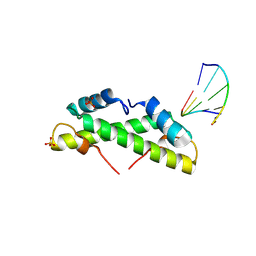 | | Bacterial histone protein HBb from Bdellovibrio bacteriovorus bound to DNA | | 分子名称: | DNA (5'-D(P*AP*GP*CP*CP*C)-3'), DNA (5'-D(P*GP*GP*GP*CP*T)-3'), PHOSPHATE ION, ... | | 著者 | Hu, Y, Albrecht, R, Hartmann, M.D. | | 登録日 | 2024-04-14 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Bacterial histone HBb from Bdellovibrio bacteriovorus compacts DNA by bending.
Nucleic Acids Res., 52, 2024
|
|
9F0E
 
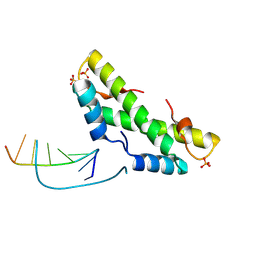 | |
8CMP
 
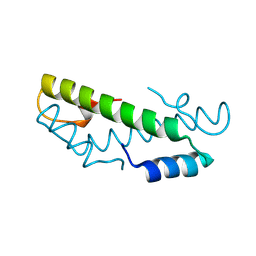 | |
2K2V
 
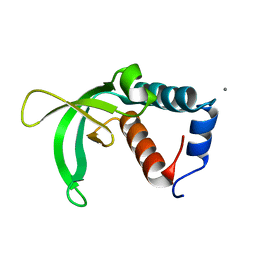 | |
2L16
 
 | |
7UPN
 
 | |
7XBG
 
 | | The crystal structure of RshSTT182/200 RBD-insert2-T346R-Y496G mutant in complex with human ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Hu, Y, Liu, K.F, Han, P, Qi, J.X. | | 登録日 | 2022-03-21 | | 公開日 | 2023-01-11 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (3.37 Å) | | 主引用文献 | Host range and structural analysis of bat-origin RshSTT182/200 coronavirus binding to human ACE2 and its animal orthologs.
Embo J., 42, 2023
|
|
7XBH
 
 | | The complex structure of RshSTT182/200 RBD bound to human ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, RshSTT182/200 coronavirus receptor binding domain, ... | | 著者 | Hu, Y, Liu, K.F, Han, P, Qi, J.X. | | 登録日 | 2022-03-21 | | 公開日 | 2023-01-11 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (3.02 Å) | | 主引用文献 | Host range and structural analysis of bat-origin RshSTT182/200 coronavirus binding to human ACE2 and its animal orthologs.
Embo J., 42, 2023
|
|
7XBF
 
 | | The complex structure of RshSTT182/200 RBD-insert2 bound to human ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, RshSTT182/200 coronavirus receptor binding domain insert2, ... | | 著者 | Hu, Y, Liu, K.F, Han, P, Qi, J.X. | | 登録日 | 2022-03-21 | | 公開日 | 2023-01-11 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (3.51 Å) | | 主引用文献 | Host range and structural analysis of bat-origin RshSTT182/200 coronavirus binding to human ACE2 and its animal orthologs.
Embo J., 42, 2023
|
|
6AGY
 
 | | Aspergillus fumigatus Af293 NDK | | 分子名称: | Nucleoside diphosphate kinase | | 著者 | Hu, Y, Han, L. | | 登録日 | 2018-08-15 | | 公開日 | 2019-03-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Characterization of crystal structure and key residues of Aspergillus fumigatus nucleoside diphosphate kinase.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
8D4L
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) S144A Mutant | | 分子名称: | 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-02 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
8D4K
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) H172Y Mutant in Complex with Inhibitor GC376 | | 分子名称: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-02 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
8D4M
 
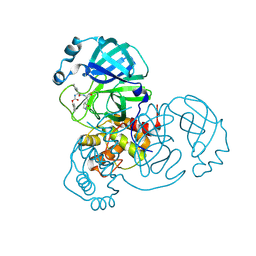 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) S144A Mutant in Complex with Inhibitor GC376 | | 分子名称: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-02 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
8D4N
 
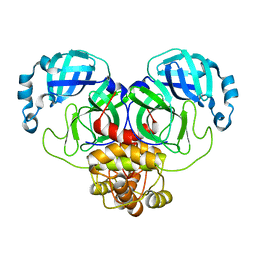 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) E166Q Mutant | | 分子名称: | 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-02 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
8D4J
 
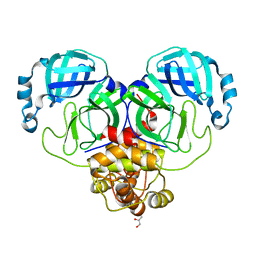 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) H172Y Mutant | | 分子名称: | 3C-like proteinase nsp5, GLYCEROL | | 著者 | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-02 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
8DZI
 
 | |
8DZH
 
 | |
