7YU0
 
 | |
7YU2
 
 | |
2N37
 
 | | Solution structure of AVR-Pia | | 分子名称: | AVR-Pia protein | | 著者 | Ose, T, Oikawa, A, Nakamura, Y, Maenaka, K, Higuchi, Y, Satoh, Y, Fujiwara, S, Demura, M, Sone, T. | | 登録日 | 2015-05-25 | | 公開日 | 2015-10-14 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of an avirulence protein, AVR-Pia, from Magnaporthe oryzae
J.Biomol.Nmr, 63, 2015
|
|
6GHP
 
 | | 14-3-3sigma in complex with a TASK3 peptide stabilized by semi-synthetic natural product FC-NAc | | 分子名称: | 14-3-3 protein sigma, CHLORIDE ION, Potassium channel subfamily K member 9, ... | | 著者 | Andrei, S.A, de Vink, P.J, Brunsveld, L, Ottmann, C, Higuchi, Y. | | 登録日 | 2018-05-08 | | 公開日 | 2018-08-01 | | 最終更新日 | 2018-10-17 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Rationally Designed Semisynthetic Natural Product Analogues for Stabilization of 14-3-3 Protein-Protein Interactions.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
1NBB
 
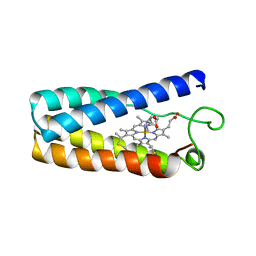 | | N-BUTYLISOCYANIDE BOUND RHODOBACTER CAPSULATUS CYTOCHROME C' | | 分子名称: | CYTOCHROME C', N-BUTYL ISOCYANIDE, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Tahirov, T.H, Misaki, S, Meyer, T.E, Cusanovich, M.A, Higuchi, Y, Yasuoka, N. | | 登録日 | 1996-03-18 | | 公開日 | 1996-08-17 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Concerted movement of side chains in the haem vicinity observed on ligand binding in cytochrome c' from rhodobacter capsulatus.
Nat.Struct.Biol., 3, 1996
|
|
3KLJ
 
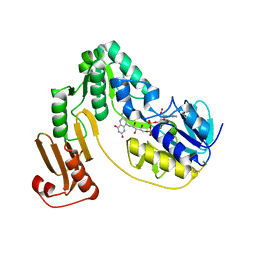 | | Crystal structure of NADH:rubredoxin oxidoreductase from Clostridium acetobutylicum | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, NAD(FAD)-dependent dehydrogenase, NirB-family (N-terminal domain) | | 著者 | Nishikawa, K, Shomura, Y, Kawasaki, S, Niimura, Y, Higuchi, Y. | | 登録日 | 2009-11-08 | | 公開日 | 2010-02-16 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of NADH:rubredoxin oxidoreductase from Clostridium acetobutylicum: a key component of the dioxygen scavenging system in obligatory anaerobes.
Proteins, 78, 2010
|
|
1QGI
 
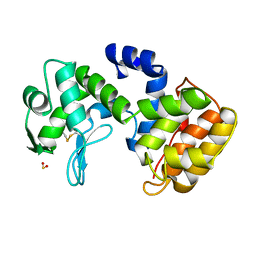 | | CHITOSANASE FROM BACILLUS CIRCULANS | | 分子名称: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (CHITOSANASE), SULFATE ION | | 著者 | Saito, J, Kita, A, Higuchi, Y, Nagata, Y, Ando, A, Miki, K. | | 登録日 | 1999-04-28 | | 公開日 | 1999-10-25 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of chitosanase from Bacillus circulans MH-K1 at 1.6-A resolution and its substrate recognition mechanism.
J.Biol.Chem., 274, 1999
|
|
1C53
 
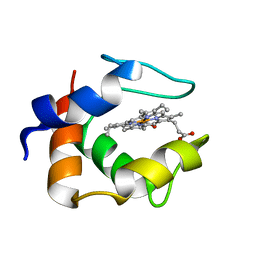 | | S-CLASS CYTOCHROMES C HAVE A VARIETY OF FOLDING PATTERNS: STRUCTURE OF CYTOCHROME C-553 FROM DESULFOVIBRIO VULGARIS DETERMINED BY THE MULTI-WAVELENGTH ANOMALOUS DISPERSION METHOD | | 分子名称: | CYTOCHROME C553, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Nakagawa, A, Higuchi, Y, Yasuoka, N, Katsube, Y, Yaga, T. | | 登録日 | 1991-08-26 | | 公開日 | 1993-10-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | S-class cytochromes c have a variety of folding patterns: structure of cytochrome c-553 from Desulfovibrio vulgaris determined by the multi-wavelength anomalous dispersion method.
J.Biochem.(Tokyo), 108, 1990
|
|
3VWL
 
 | | Crystal structure of 6-aminohexanoate-dimer hydrolase G181D/R187S/H266N/D370Y mutant | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | 著者 | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | 登録日 | 2012-08-30 | | 公開日 | 2013-10-16 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
To be Published
|
|
3VWR
 
 | | Crystal structure of 6-aminohexanoate-dimer hydrolase S112A/G181D/R187G/H266N/D370Y mutant complexd with 6-aminohexanoate | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | 著者 | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | 登録日 | 2012-08-30 | | 公開日 | 2013-10-16 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
To be Published
|
|
3VWM
 
 | | Crystal structure of 6-aminohexanoate-dimer hydrolase G181D/R187A/H266N/D370Y mutant | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | 著者 | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | 登録日 | 2012-08-30 | | 公開日 | 2013-10-16 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
To be Published
|
|
1FLM
 
 | | DIMER OF FMN-BINDING PROTEIN FROM DESULFOVIBRIO VULGARIS (MIYAZAKI F) | | 分子名称: | FLAVIN MONONUCLEOTIDE, PROTEIN (FMN-BINDING PROTEIN) | | 著者 | Suto, K, Kawagoe, K, Shibata, N, Morimoto, K, Higuchi, Y, Kitamura, M, Nakaya, T, Yasuoka, N. | | 登録日 | 1999-03-10 | | 公開日 | 2000-03-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | How do the x-ray structure and the NMR structure of FMN-binding protein differ?
Acta Crystallogr.,Sect.D, 56, 2000
|
|
6L1V
 
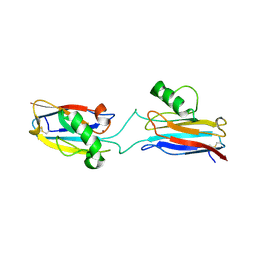 | | Domain-swapped Alcaligenes xylosoxidans azurin dimer | | 分子名称: | Azurin-1, COPPER (II) ION | | 著者 | Cahyono, R.N, Yamanaka, M, Nagao, S, Shibata, N, Higuchi, Y, Hirota, S. | | 登録日 | 2019-09-30 | | 公開日 | 2020-02-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | 3D domain swapping of azurin from Alcaligenes xylosoxidans.
Metallomics, 12, 2020
|
|
6LS8
 
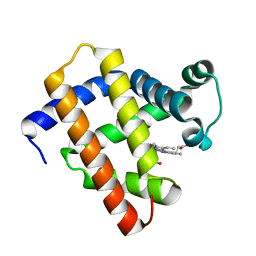 | | The monomeric structure of G80A/H81A/H82A myoglobin | | 分子名称: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Nagao, S, Suda, A, Kobayashi, H, Shibata, N, Higuchi, Y, Hirota, S. | | 登録日 | 2020-01-17 | | 公開日 | 2020-05-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Thermodynamic Control of Domain Swapping by Modulating the Helical Propensity in the Hinge Region of Myoglobin.
Chem Asian J, 15, 2020
|
|
6LTM
 
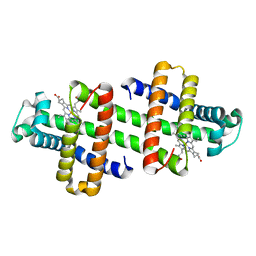 | | The dimeric structure of G80A/H81A/H82A myoglobin | | 分子名称: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Nagao, S, Suda, A, Kobayashi, H, Shibata, N, Higuchi, Y, Hirota, S. | | 登録日 | 2020-01-22 | | 公開日 | 2020-05-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Thermodynamic Control of Domain Swapping by Modulating the Helical Propensity in the Hinge Region of Myoglobin.
Chem Asian J, 15, 2020
|
|
4P7X
 
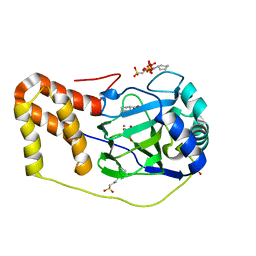 | | L-pipecolic acid-bound L-proline cis-4-hydroxylase | | 分子名称: | (2S)-piperidine-2-carboxylic acid, 2-OXOGLUTARIC ACID, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ... | | 著者 | Shomura, Y, Koketsu, K, Moriwaki, K, Hayashi, M, Mitsuhashi, S, Hara, R, Kino, K, Higuchi, Y. | | 登録日 | 2014-03-28 | | 公開日 | 2014-09-17 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Refined Regio- and Stereoselective Hydroxylation of l-Pipecolic Acid by Protein Engineering of l-Proline cis-4-Hydroxylase Based on the X-ray Crystal Structure.
Acs Synth Biol, 4, 2015
|
|
3L3F
 
 | |
1J0P
 
 | | Three dimensional Structure of the Y43L mutant of Tetraheme Cytochrome c3 from Desulfovibrio vulgaris Miyazaki F | | 分子名称: | Cytochrome c3, ETHANOL, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Ozawa, K, Yasukawa, F, Kumagai, J, Ohmura, T, Cusanvich, M.A, Tomimoto, Y, Ogata, H, Higuchi, Y, Akutsu, H. | | 登録日 | 2002-11-19 | | 公開日 | 2003-11-19 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (0.91 Å) | | 主引用文献 | Role of the aromatic ring of Tyr43 in tetraheme cytochrome c(3) from Desulfovibrio vulgaris Miyazaki F.
Biophys.J., 85, 2003
|
|
1J0O
 
 | | High Resolution Crystal Structure of the wild type Tetraheme Cytochrome c3 from Desulfovibrio vulgaris Miyazaki F | | 分子名称: | Cytochrome c3, ETHANOL, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Ozawa, K, Yasukawa, F, Kumagai, J, Ohmura, T, Cusanvich, M.A, Tomimoto, Y, Ogata, H, Higuchi, Y, Akutsu, H. | | 登録日 | 2002-11-19 | | 公開日 | 2003-11-19 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Role of the aromatic ring of Tyr43 in tetraheme cytochrome c(3) from Desulfovibrio vulgaris Miyazaki F.
Biophys.J., 85, 2003
|
|
3VMM
 
 | |
8HGM
 
 | | Structure of SARS-CoV-2 spike RBD in complex with neutralizing antibody NIV-11 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-11 Fab heavy chain, NIV-11 Fab light chain, ... | | 著者 | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | 登録日 | 2022-11-15 | | 公開日 | 2023-10-25 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
8HGL
 
 | | SARS-CoV-2 spike in complex with neutralizing antibody NIV-11 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-11 Fab heavy chain, ... | | 著者 | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | 登録日 | 2022-11-15 | | 公開日 | 2023-10-25 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
8HES
 
 | | Crystal structure of SARS-CoV-2 RBD and NIV-10 complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-10 Fab H-chain, NIV-10 Fab L-chain, ... | | 著者 | Moriyama, S, Anraku, Y, Taminishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | 登録日 | 2022-11-08 | | 公開日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
4HAL
 
 | | Multicopper Oxidase CueO mutant E506I | | 分子名称: | ACETATE ION, Blue copper oxidase CueO, COPPER (II) ION, ... | | 著者 | Komori, H, Kataoka, K, Sakurai, T, Higuchi, Y. | | 登録日 | 2012-09-26 | | 公開日 | 2013-10-30 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Multicopper Oxidase CueO mutant E506I
TO BE PUBLISHED
|
|
4HAK
 
 | | Multicopper Oxidase CueO mutant E506A | | 分子名称: | ACETATE ION, Blue copper oxidase CueO, COPPER (II) ION, ... | | 著者 | Komori, H, Kataoka, K, Sakurai, T, Higuchi, Y. | | 登録日 | 2012-09-26 | | 公開日 | 2013-10-30 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Multicopper Oxidase CueO mutant E506A
TO BE PUBLISHED
|
|
