7P7W
 
 | | N-acetylglucosamine kinase from Plesiomonas shigelloides compexed with alpha-N-acetylglucosamine and ADP | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | 登録日 | 2021-07-20 | | 公開日 | 2022-08-03 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
7P7I
 
 | | Native structure of N-acetylglucosamine kinase from Plesiomonas shigelloides | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | 著者 | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | 登録日 | 2021-07-19 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
7P9L
 
 | | N-acetylglucosamine kinase from Plesiomonas shigelloides compexed with alpha-N-acetylglucosamine-6-phosphate | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-6-O-phosphono-beta-D-glucopyranose, ... | | 著者 | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | 登録日 | 2021-07-27 | | 公開日 | 2022-08-10 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
7P9P
 
 | | N-acetylglucosamine kinase from Plesiomonas shigelloides compexed with alpha-N-acetylglucosamine and AMP-PNP inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | 登録日 | 2021-07-27 | | 公開日 | 2022-08-10 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
7PA1
 
 | | Structure of N-acetylglucosamine kinase from Plesiomonas shigelloides in complex with AMP-PNP in the absence of N-acetylglucoseamine substrate | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | 登録日 | 2021-07-28 | | 公開日 | 2022-08-10 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
7P9Y
 
 | | N-acetylglucosamine kinase from Plesiomonas shigelloides compexed with alpha-N-acetylglucosamine | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, ... | | 著者 | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | 登録日 | 2021-07-28 | | 公開日 | 2022-08-10 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
5LU5
 
 | | A quantum half-site enzyme | | 分子名称: | 7-O-phosphono-D-glycero-alpha-D-manno-heptopyranose, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Vivoli, M, Harmer, N.J, Pang, J. | | 登録日 | 2016-09-08 | | 公開日 | 2017-12-06 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | A half-site multimeric enzyme achieves its cooperativity without conformational changes.
Sci Rep, 7, 2017
|
|
5LTZ
 
 | | GmhA_mutant Q175E | | 分子名称: | 1,2-ETHANEDIOL, D-ALTRO-HEPT-2-ULOSE 7-PHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Vivoli, M, Harmer, N.J. | | 登録日 | 2016-09-07 | | 公開日 | 2017-12-06 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | A half-site multimeric enzyme achieves its cooperativity without conformational changes.
Sci Rep, 7, 2017
|
|
5LU6
 
 | | Heptose isomerase mutant - H64Q | | 分子名称: | 1,2-ETHANEDIOL, D-ALTRO-HEPT-2-ULOSE 7-PHOSPHATE, GLYCEROL, ... | | 著者 | Vivoli, M, Harmer, N.J, Pang, J. | | 登録日 | 2016-09-08 | | 公開日 | 2017-12-06 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | A half-site multimeric enzyme achieves its cooperativity without conformational changes.
Sci Rep, 7, 2017
|
|
5LU7
 
 | | Heptose isomerase GmhA mutant - D61A | | 分子名称: | 1,2-ETHANEDIOL, 7-O-phosphono-D-glycero-alpha-D-manno-heptopyranose, Phosphoheptose isomerase, ... | | 著者 | Vivoli, M, Harmer, N.J, Pang, J. | | 登録日 | 2016-09-08 | | 公開日 | 2017-12-06 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | A half-site multimeric enzyme achieves its cooperativity without conformational changes.
Sci Rep, 7, 2017
|
|
4BQO
 
 | | Structural insights into WcbI, a novel polysaccharide biosynthesis enzyme. Native protein without disulfide bond between COA and Cys14. | | 分子名称: | BROMIDE ION, COENZYME A, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Vivoli, M, Ayres, E, Isupov, M.N, Harmer, N.J. | | 登録日 | 2013-05-31 | | 公開日 | 2013-11-06 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Structural Insights Into Wcbi, a Novel Polysaccharide-Biosynthesis Enzyme.
Iucrj, 1, 2014
|
|
4BQN
 
 | | Structural insights into WcbI, a novel polysaccharide biosynthesis enzyme. Native protein. | | 分子名称: | CAPSULAR POLYSACCHARIDE BIOSYNTHESIS PROTEIN, CHLORIDE ION, COENZYME A, ... | | 著者 | Vivoli, M, Ayres, E, Isupov, M.N, Harmer, N.J. | | 登録日 | 2013-05-31 | | 公開日 | 2013-11-06 | | 最終更新日 | 2014-08-13 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Structural Insights Into Wcbi, a Novel Polysaccharide-Biosynthesis Enzyme.
Iucrj, 1, 2014
|
|
7PQ9
 
 | | Crystal structure of Bacillus clausii pdxR at 2.8 Angstroms resolution | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Vivoli Vega, M, Isupov, M.N, Harmer, N. | | 登録日 | 2021-09-16 | | 公開日 | 2022-09-28 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
8ORU
 
 | | cyclic 2,3-diphosphoglycerate synthetase from the hyperthermophilic archaeon Methanothermus fervidus bound to 2,3-diphosphoglycerate and ADP. | | 分子名称: | (2R)-2,3-diphosphoglyceric acid, 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | De Rose, S.A, Isupov, M. | | 登録日 | 2023-04-17 | | 公開日 | 2023-12-06 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Structural characterization of a novel cyclic 2,3-diphosphoglycerate synthetase involved in extremolyte production in the archaeon Methanothermus fervidus .
Front Microbiol, 14, 2023
|
|
8ORK
 
 | | cyclic 2,3-diphosphoglycerate synthetase from the hyperthermophilic archaeon Methanothermus fervidus | | 分子名称: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | 著者 | De Rose, S.A, Isupov, M. | | 登録日 | 2023-04-14 | | 公開日 | 2023-12-06 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Structural characterization of a novel cyclic 2,3-diphosphoglycerate synthetase involved in extremolyte production in the archaeon Methanothermus fervidus .
Front Microbiol, 14, 2023
|
|
6O49
 
 | |
6O4A
 
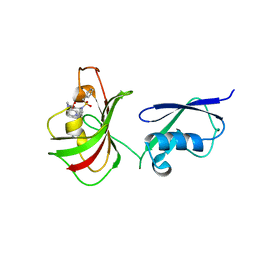 | |
7ZTH
 
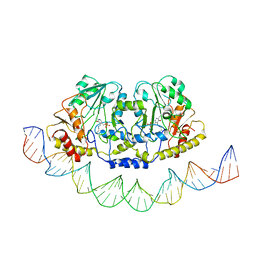 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the open conformation | | 分子名称: | DNA (48-MER), PLP-dependent aminotransferase family protein | | 著者 | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | 登録日 | 2022-05-10 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZN5
 
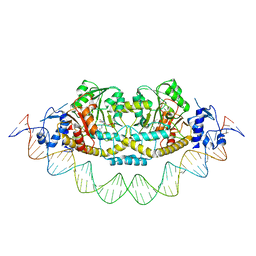 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the closed conformation, C2 symmetry. | | 分子名称: | DNA (48-MER), PLP-dependent aminotransferase family protein | | 著者 | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | 登録日 | 2022-04-20 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZLA
 
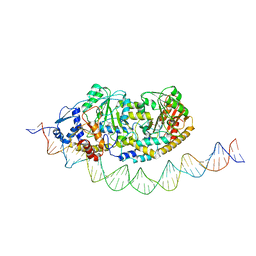 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the half-closed conformation | | 分子名称: | DNA (48-MER), PLP-dependent aminotransferase family protein | | 著者 | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Savino, C, Exertier, C, Bolognesi, M, Chaves Sanjuan, A. | | 登録日 | 2022-04-14 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (3.99 Å) | | 主引用文献 | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZPA
 
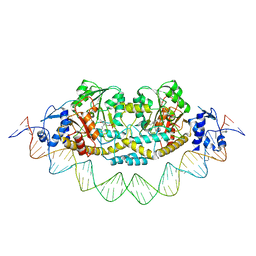 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the closed conformation, C1 symmetry | | 分子名称: | DNA (48-MER), PLP-dependent aminotransferase family protein | | 著者 | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | 登録日 | 2022-04-27 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
5FRD
 
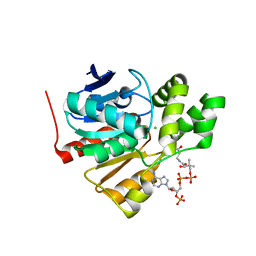 | | Structure of a thermophilic esterase | | 分子名称: | CARBOXYLESTERASE (EST-2), CHLORIDE ION, CITRATE ANION, ... | | 著者 | Sayer, C, Finnigan, W, Isupov, M.N, Levisson, M, Kengen, S.W.M, van der Oost, J, Harmer, N, Littlechild, J.A. | | 登録日 | 2015-12-17 | | 公開日 | 2016-05-25 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural and Biochemical Characterisation of Archaeoglobus Fulgidus Esterase Reveals a Bound Coa Molecule in the Vicinity of the Active Site.
Sci.Rep., 6, 2016
|
|
6G1C
 
 | |
6G1N
 
 | |
6G26
 
 | | The crystal structure of the Burkholderia pseudomallei HicAB complex | | 分子名称: | 1,2-ETHANEDIOL, HicA, HicB, ... | | 著者 | Winter, A.J, Isupov, M.N, Williams, C, Crump, M.P. | | 登録日 | 2018-03-22 | | 公開日 | 2018-10-31 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | The molecular basis of protein toxin HicA-dependent binding of the protein antitoxin HicB to DNA.
J. Biol. Chem., 293, 2018
|
|
