2VFO
 
 | | Low Temperature Structure of P22 Tailspike Protein Fragment (109-666), Mutant V125L | | 分子名称: | CALCIUM ION, GLYCEROL, P22 TAILSPIKE PROTEIN, ... | | 著者 | Becker, M, Mueller, J.J, Heinemann, U, Seckler, R. | | 登録日 | 2007-11-05 | | 公開日 | 2008-12-16 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Side-Chain Stacking and Beta-Helix Stability in P22 Tailspike Protein
To be Published
|
|
2VFM
 
 | | Low Temperature Structure of P22 Tailspike Protein Fragment (109-666) | | 分子名称: | BIFUNCTIONAL TAIL PROTEIN, CALCIUM ION, GLYCEROL, ... | | 著者 | Becker, M, Mueller, J.J, Heinemann, U, Seckler, R. | | 登録日 | 2007-11-05 | | 公開日 | 2008-12-16 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Side-Chain Stacking and Beta-Helix Stability in P22 Tailspike Protein
To be Published
|
|
2VFQ
 
 | | Low Temperature Structure of P22 Tailspike Protein Fragment (109-666), Mutant V450A | | 分子名称: | CALCIUM ION, GLYCEROL, P22 TAILSPIKE PROTEIN,, ... | | 著者 | Becker, M, Mueller, J.J, Heinemann, U, Seckler, R. | | 登録日 | 2007-11-05 | | 公開日 | 2008-12-16 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Side-Chain Stacking and Beta-Helix Stability in P22 Tailspike Protein
To be Published
|
|
2VFN
 
 | | Low Temperature Structure of P22 Tailspike Protein Fragment (109-666), Mutant V125A | | 分子名称: | BIFUNCTIONAL TAIL PROTEIN, CALCIUM ION, GLYCEROL, ... | | 著者 | Becker, M, Mueller, J.J, Heinemann, U, Seckler, R. | | 登録日 | 2007-11-05 | | 公開日 | 2008-12-16 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Side-Chain Stacking and Beta-Helix Stability in P22 Tailspike Protein
To be Published
|
|
2VFP
 
 | | Low Temperature Structure of P22 Tailspike Protein Fragment (109-666), Mutant V349L | | 分子名称: | CALCIUM ION, GLYCEROL, P22 TAILSPIKE PROTEIN, ... | | 著者 | Becker, M, Mueller, J.J, Heinemann, U, Seckler, R. | | 登録日 | 2007-11-05 | | 公開日 | 2008-12-16 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Side-Chain Stacking and Beta-Helix Stability in P22 Tailspike Protein
To be Published
|
|
4JIN
 
 | | X-ray crystal structure of Archaeoglobus fulgidus Rio1 bound to (2E)-N-benzyl-2-cyano-3-(pyridine-4-yl)acrylamide (WP1086) | | 分子名称: | (2E)-N-benzyl-2-cyano-3-(pyridin-4-yl)prop-2-enamide, RIO-type serine/threonine-protein kinase Rio1 | | 著者 | Mielecki, M, Krawiec, K, Kiburu, I, Grzelak, K, Wlodzimierz, Z, Kierdaszuk, B, Kowa, K, Fokt, I, Szymanski, S, Piotr, S, Szeja, W, Priebe, W, Lesyng, B, LaRonde-LeBlanc, N. | | 登録日 | 2013-03-06 | | 公開日 | 2013-04-24 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.095 Å) | | 主引用文献 | Development of novel molecular probes of the Rio1 atypical protein kinase.
Biochim.Biophys.Acta, 1834, 2013
|
|
6NUX
 
 | | CD1a-lipid binary complex | | 分子名称: | (2E,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol, 1,2-ETHANEDIOL, Beta-2-microglobulin, ... | | 著者 | Wegrecki, M, Le Nours, J, Rossjohn, J. | | 登録日 | 2019-02-03 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Human T cell response to CD1a and contact dermatitis allergens in botanical extracts and commercial skin care products.
Sci Immunol, 5, 2020
|
|
6R2H
 
 | | Crystal structure of Apo PinO from Porphyromonas gingivitis | | 分子名称: | GLYCEROL, HmuY protein | | 著者 | Antonyuk, S.V, Bielecki, M, Strange, R.W, Capper, M, Olczak, T, Olczak, M. | | 登録日 | 2019-03-17 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | Prevotella intermedia produces two proteins homologous to Porphyromonas gingivalis HmuY but with different heme coordination mode.
Biochem.J., 477, 2020
|
|
8OYT
 
 | | Stabilised BA.1 SARS-CoV-2 spike with H6 nanobodies in '3 up' RBD conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, H6 nanobody, ... | | 著者 | Weckener, M, Naismith, J.H, Owens, R.J. | | 登録日 | 2023-05-05 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural and functional characterization of nanobodies that neutralize Omicron variants of SARS-CoV-2.
Open Biology, 14, 2024
|
|
8OYU
 
 | | Stabilised BA.1 SARS-CoV-2 spike with H6 nanobodies in '2 up 1 down' RBD conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, H6 nanobody, ... | | 著者 | Weckener, M, Naismith, J.H, Owens, R.J. | | 登録日 | 2023-05-05 | | 公開日 | 2024-05-15 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structural and functional characterization of nanobodies that neutralize Omicron variants of SARS-CoV-2.
Open Biology, 14, 2024
|
|
2N64
 
 | |
6EWM
 
 | | Crystal structure of heme free PORPHYROMONAS GINGIVALIS HEME-BINDING PROTEIN HMUY | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Haemophore HmuY, ... | | 著者 | Antonyuk, S.V, Strange, R.W, Bielecki, M, Olczak, T, Olczak, M. | | 登録日 | 2017-11-05 | | 公開日 | 2018-10-24 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Tannerella forsythiaTfo belongs toPorphyromonas gingivalisHmuY-like family of proteins but differs in heme-binding properties.
Biosci. Rep., 38, 2018
|
|
7SH4
 
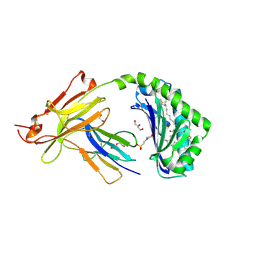 | | CD1a-phosphatidylglycerol binary structure | | 分子名称: | (21R,24R,27S)-24,27,28-trihydroxy-18,24-dioxo-19,23,25-trioxa-24lambda~5~-phosphaoctacosan-21-yl (9Z)-octadec-9-enoate, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | 著者 | Wegrecki, M, Rossjohn, J. | | 登録日 | 2021-10-07 | | 公開日 | 2022-10-05 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Staphylococcal phosphatidylglycerol antigens activate human T cells via CD1a.
Nat.Immunol., 24, 2023
|
|
9DNW
 
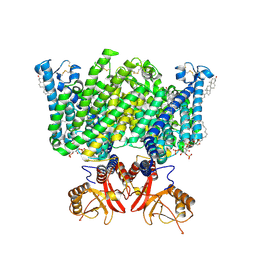 | | Human ClC-3:noTMEM9 | | 分子名称: | (2R)-1-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,4,6-trihydroxy-3,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5E,8E,11E,13E)-icosa-5,8,11,13-tetraenoate, CHLORIDE ION, CHOLESTEROL, ... | | 著者 | Son, Y, Schrecker, M, Hite, R.K. | | 登録日 | 2024-09-18 | | 公開日 | 2025-04-02 | | 最終更新日 | 2025-05-28 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis of ClC-3 inhibition by TMEM9 and PI(3,5)P 2.
Biorxiv, 2025
|
|
9DNZ
 
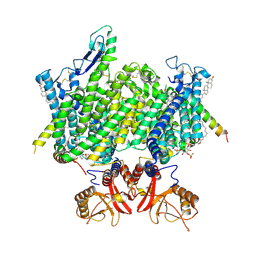 | | Human ClC-3:TMEM9, TMEM9 Protomer A: Complete, TMEM9 Protomer B: No LD, No CD | | 分子名称: | (2R)-1-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,4,6-trihydroxy-3,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5E,8E,11E,13E)-icosa-5,8,11,13-tetraenoate, CHLORIDE ION, CHOLESTEROL, ... | | 著者 | Son, Y, Schrecker, M, Hite, R.K. | | 登録日 | 2024-09-18 | | 公開日 | 2025-04-02 | | 最終更新日 | 2025-05-21 | | 実験手法 | ELECTRON MICROSCOPY (3.16 Å) | | 主引用文献 | Structural basis of ClC-3 inhibition by TMEM9 and PI(3,5)P 2.
Biorxiv, 2025
|
|
9DNX
 
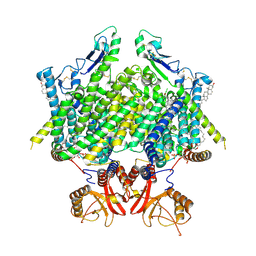 | | Human ClC-3:TMEM9, TMEM9 Protomer A and B: Complete | | 分子名称: | (2R)-1-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,4,6-trihydroxy-3,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5E,8E,11E,13E)-icosa-5,8,11,13-tetraenoate, CHLORIDE ION, CHOLESTEROL, ... | | 著者 | Son, Y, Schrecker, M, Hite, R.K. | | 登録日 | 2024-09-18 | | 公開日 | 2025-04-02 | | 最終更新日 | 2025-05-28 | | 実験手法 | ELECTRON MICROSCOPY (2.86 Å) | | 主引用文献 | Structural basis of ClC-3 inhibition by TMEM9 and PI(3,5)P 2.
Biorxiv, 2025
|
|
9DNY
 
 | | Human ClC-3:TMEM9, TMEM9 Protomer A: No CD TMEM9, Protomer B: No LD, No CD | | 分子名称: | (2R)-1-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,4,6-trihydroxy-3,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5E,8E,11E,13E)-icosa-5,8,11,13-tetraenoate, CHLORIDE ION, CHOLESTEROL, ... | | 著者 | Son, Y, Schrecker, M, Hite, R.K. | | 登録日 | 2024-09-18 | | 公開日 | 2025-04-02 | | 最終更新日 | 2025-05-28 | | 実験手法 | ELECTRON MICROSCOPY (3.01 Å) | | 主引用文献 | Structural basis of ClC-3 inhibition by TMEM9 and PI(3,5)P 2.
Biorxiv, 2025
|
|
6Z1F
 
 | | CryoEM structure of Rubisco Activase with its substrate Rubisco from Nostoc sp. (strain PCC7120) | | 分子名称: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Wang, H, Bracher, A, Flecken, M, Popilka, L, Hartl, F.U, Hayer-Hartl, M. | | 登録日 | 2020-05-13 | | 公開日 | 2020-09-23 | | 最終更新日 | 2025-04-09 | | 実験手法 | ELECTRON MICROSCOPY (2.86 Å) | | 主引用文献 | Dual Functions of a Rubisco Activase in Metabolic Repair and Recruitment to Carboxysomes.
Cell, 183, 2020
|
|
6Z1G
 
 | | CryoEM structure of the interaction between Rubisco Activase small-subunit-like (SSUL) domain with Rubisco from Nostoc sp. (strain PCC7120) | | 分子名称: | Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase small chain, Ribulose bisphosphate carboxylase/oxygenase activase | | 著者 | Wang, H, Bracher, A, Flecken, M, Popilka, L, Hartl, F.U, Hayer-Hartl, M. | | 登録日 | 2020-05-13 | | 公開日 | 2020-09-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (8.2 Å) | | 主引用文献 | Dual Functions of a Rubisco Activase in Metabolic Repair and Recruitment to Carboxysomes.
Cell, 183, 2020
|
|
7Z86
 
 | |
7Z6V
 
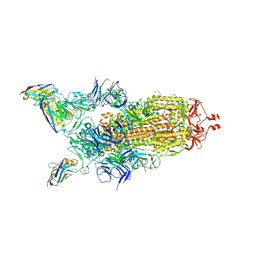 | | CRYO-EM STRUCTURE OF SARS-COV-2 SPIKE : H11 nanobody complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody H11, ... | | 著者 | Weckener, M, Naismith, J.H, Vogirala, V.K. | | 登録日 | 2022-03-14 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z85
 
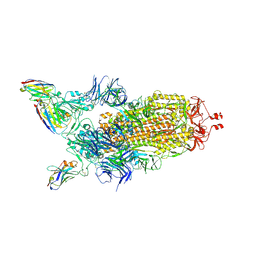 | | CRYO-EM STRUCTURE OF SARS-COV-2 SPIKE : H11-B5 nanobody complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody H11-B5, ... | | 著者 | Weckener, M, Naismith, J.H. | | 登録日 | 2022-03-16 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z9Q
 
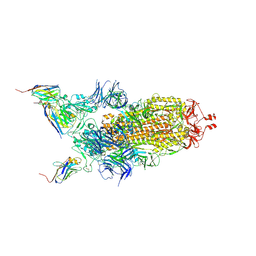 | | CRYO-EM STRUCTURE OF SARS-COV-2 SPIKE : H11-A10 nanobody complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody H11-A10, ... | | 著者 | Weckener, M, Naismith, J.H. | | 登録日 | 2022-03-21 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z7X
 
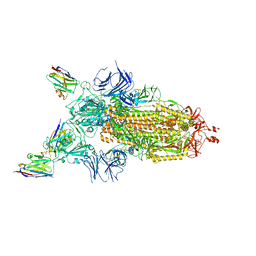 | | CRYO-EM STRUCTURE OF SARS-COV-2 SPIKE : H11-H6 nanobody complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody H11-H6, ... | | 著者 | Weckener, M, Naismith, J.H. | | 登録日 | 2022-03-16 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z9R
 
 | |
