3VSK
 
 | |
3VSL
 
 | | Crystal structure of penicillin-binding protein 3 (PBP3) from methicilin-resistant Staphylococcus aureus in the cefotaxime bound form. | | 分子名称: | CEFOTAXIME, C3' cleaved, open, ... | | 著者 | Yoshida, H, Tame, J.R, Park, S.Y. | | 登録日 | 2012-04-25 | | 公開日 | 2012-10-31 | | 最終更新日 | 2017-03-15 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structures of Penicillin-Binding Protein 3 (PBP3) from Methicillin-Resistant Staphylococcus aureus in the Apo and Cefotaxime-Bound Forms.
J.Mol.Biol., 423, 2012
|
|
5Z01
 
 | |
6AL1
 
 | | The NZ-1 Fab complexed with the PDZ tandem fragment of A. aeolicus S2P homolog with the PA12 tag inserted between the residues 181 and 184 | | 分子名称: | Heavy chain of antigen binding fragment, Fab of NZ-1, Light chain of antigen binding fragment, ... | | 著者 | Tamura, R, Oi, R, Kaneko, M.K, Kato, Y, Nogi, T. | | 登録日 | 2018-09-05 | | 公開日 | 2019-02-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Application of the NZ-1 Fab as a crystallization chaperone for PA tag-inserted target proteins.
Protein Sci., 28, 2019
|
|
6AKQ
 
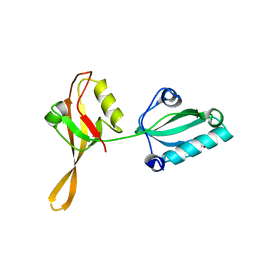 | | The PDZ tandem fragment of A. aeolicus S2P homolog with the PA12 tag inserted between the residues 263 and 267 | | 分子名称: | PDZ tandem fragment of A. aeolicus site-2 protease homolog with the PA tag insertion | | 著者 | Tamura, R, Oi, R, Kaneko, M.K, Kato, Y, Nogi, T. | | 登録日 | 2018-09-03 | | 公開日 | 2019-02-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Application of the NZ-1 Fab as a crystallization chaperone for PA tag-inserted target proteins.
Protein Sci., 28, 2019
|
|
6AL0
 
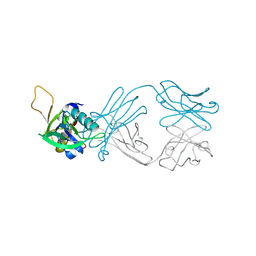 | | The NZ-1 Fab complexed with the PDZ tandem fragment of A. aeolicus S2P homolog with the PA12 tag inserted between the residues 263 and 267 | | 分子名称: | Heavy chain of antigen binding fragment, Fab of NZ-1, Light chain of antigen binding fragment, ... | | 著者 | Tamura, R, Oi, R, Kaneko, M.K, Kato, Y, Nogi, T. | | 登録日 | 2018-09-05 | | 公開日 | 2019-02-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Application of the NZ-1 Fab as a crystallization chaperone for PA tag-inserted target proteins.
Protein Sci., 28, 2019
|
|
5Z03
 
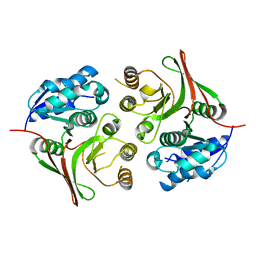 | |
6TJF
 
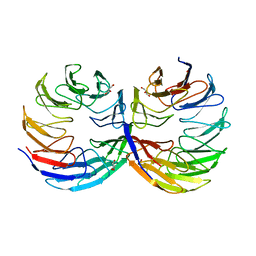 | | Crystal structure of the computationally designed Cake6 protein | | 分子名称: | Cake6, GLYCEROL | | 著者 | Mylemans, B, Laier, I, Voet, A.R.D, Noguchi, H. | | 登録日 | 2019-11-26 | | 公開日 | 2020-05-06 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural plasticity of a designer protein sheds light on beta-propeller protein evolution.
Febs J., 288, 2021
|
|
6TJE
 
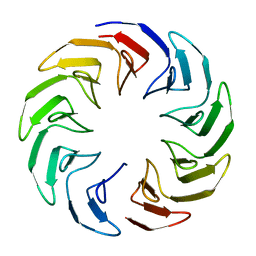 | |
6TJH
 
 | | Crystal structure of the computationally designed Cake9 protein | | 分子名称: | Cake9, GLYCEROL, SULFATE ION | | 著者 | Mylemans, B, Laier, I, Noguchi, H, Voet, A.R.D. | | 登録日 | 2019-11-26 | | 公開日 | 2020-05-06 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Structural plasticity of a designer protein sheds light on beta-propeller protein evolution.
Febs J., 288, 2021
|
|
6TJC
 
 | | Crystal structure of the computationally designed Cake3 protein | | 分子名称: | Cake3, GLYCEROL, PHOSPHATE ION | | 著者 | Laier, I, Mylemans, B, Voet, A.R.D, Noguchi, H. | | 登録日 | 2019-11-26 | | 公開日 | 2020-05-06 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural plasticity of a designer protein sheds light on beta-propeller protein evolution.
Febs J., 288, 2021
|
|
6TJG
 
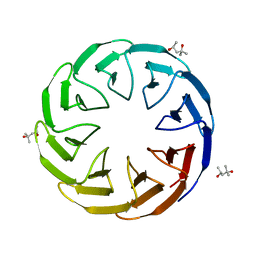 | | Crystal structure of the computationally designed Cake8 protein | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cake8 | | 著者 | Laier, I, Mylemans, B, Noguchi, H, Voet, A.R.D. | | 登録日 | 2019-11-26 | | 公開日 | 2020-05-06 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural plasticity of a designer protein sheds light on beta-propeller protein evolution.
Febs J., 288, 2021
|
|
6TJD
 
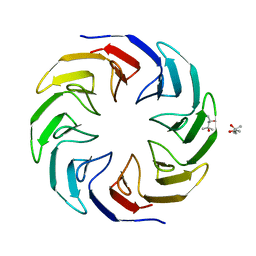 | | Crystal structure of the computationally designed Cake4 protein | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cake4 | | 著者 | Laier, I, Mylemans, B, Noguchi, H, Voet, A.R.D. | | 登録日 | 2019-11-26 | | 公開日 | 2020-05-06 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural plasticity of a designer protein sheds light on beta-propeller protein evolution.
Febs J., 288, 2021
|
|
6TJB
 
 | | Crystal structure of the computationally designed Cake2 protein | | 分子名称: | Cake2, GLYCEROL | | 著者 | Laier, I, Mylemans, B, Noguchi, H, Voet, A.R.D. | | 登録日 | 2019-11-26 | | 公開日 | 2020-05-06 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural plasticity of a designer protein sheds light on beta-propeller protein evolution.
Febs J., 288, 2021
|
|
6TJI
 
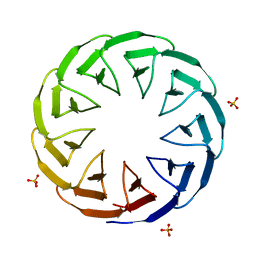 | | Crystal structure of the computationally designed Cake10 protein | | 分子名称: | Cake10, PHOSPHATE ION | | 著者 | Laier, I, Mylemans, B, Voet, A.R.D, Noguchi, H. | | 登録日 | 2019-11-26 | | 公開日 | 2020-05-06 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural plasticity of a designer protein sheds light on beta-propeller protein evolution.
Febs J., 288, 2021
|
|
7W71
 
 | |
7Y4P
 
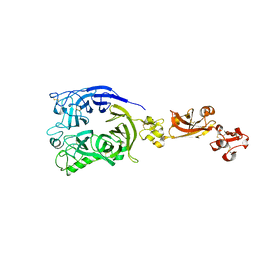 | | Human Plexin A1, extracellular domains 1-4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Plexin-A1 | | 著者 | Tanaka, T, Neyazaki, M, Nogi, T. | | 登録日 | 2022-06-15 | | 公開日 | 2022-10-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Hybrid in vitro/in silico analysis of low-affinity protein-protein interactions that regulate signal transduction by Sema6D.
Protein Sci., 31, 2022
|
|
7Y4O
 
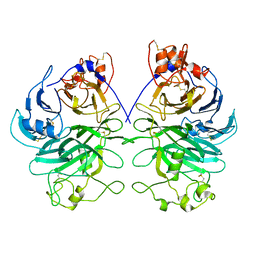 | | Rat Semaphorin 6D extracellular region | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Semaphorin 6D | | 著者 | Tanaka, T, Neyazaki, M, Nogi, T. | | 登録日 | 2022-06-15 | | 公開日 | 2022-10-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Hybrid in vitro/in silico analysis of low-affinity protein-protein interactions that regulate signal transduction by Sema6D.
Protein Sci., 31, 2022
|
|
7Y4Q
 
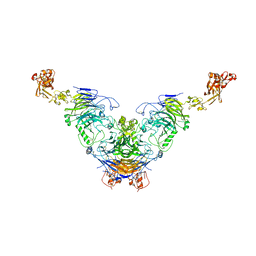 | | Semaphorin 6D in complex with Plexin A1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Plexin-A1, ... | | 著者 | Tanaka, T, Neyazaki, M, Nogi, T. | | 登録日 | 2022-06-16 | | 公開日 | 2022-10-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (4.7 Å) | | 主引用文献 | Hybrid in vitro/in silico analysis of low-affinity protein-protein interactions that regulate signal transduction by Sema6D.
Protein Sci., 31, 2022
|
|
7W6Z
 
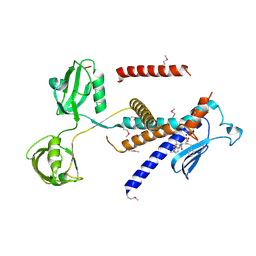 | |
7W70
 
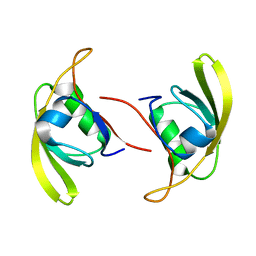 | |
7W6X
 
 | |
7W6Y
 
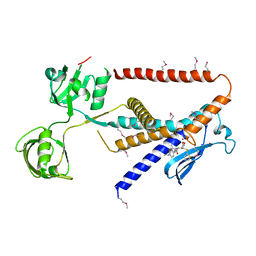 | | Crystal structure of Kangiella koreensis RseP orthologue in complex with batimastat in space group P1 | | 分子名称: | 4-(N-HYDROXYAMINO)-2R-ISOBUTYL-2S-(2-THIENYLTHIOMETHYL)SUCCINYL-L-PHENYLALANINE-N-METHYLAMIDE, Anti sigma-E protein, RseA, ... | | 著者 | Imaizumi, Y, Takanuki, K, Nogi, T. | | 登録日 | 2021-12-02 | | 公開日 | 2022-09-07 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Mechanistic insights into intramembrane proteolysis by E. coli site-2 protease homolog RseP.
Sci Adv, 8, 2022
|
|
7X58
 
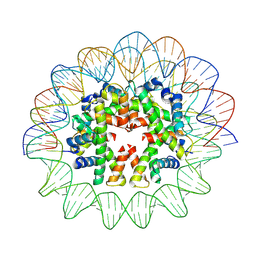 | | Cryo-EM structure of human subnucleosome (open form) | | 分子名称: | Histone H3.1, Histone H4, Widom601 DNA FW (145-MER), ... | | 著者 | Nozawa, K, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2022-03-04 | | 公開日 | 2022-11-16 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.93 Å) | | 主引用文献 | Cryo-electron microscopy structure of the H3-H4 octasome: A nucleosome-like particle without histones H2A and H2B.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7X57
 
 | | Cryo-EM structure of human subnucleosome (closed form) | | 分子名称: | Histone H3.1, Histone H4, Widom601 DNA FW (145-MER), ... | | 著者 | Nozawa, K, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2022-03-04 | | 公開日 | 2022-11-16 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.63 Å) | | 主引用文献 | Cryo-electron microscopy structure of the H3-H4 octasome: A nucleosome-like particle without histones H2A and H2B.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
