1FK2
 
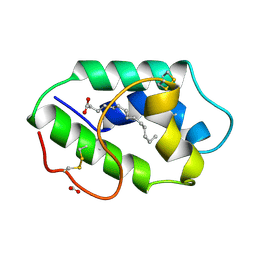 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH MYRISTIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | 分子名称: | FORMIC ACID, MYRISTIC ACID, NONSPECIFIC LIPID-TRANSFER PROTEIN | | 著者 | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | 登録日 | 2000-08-09 | | 公開日 | 2001-06-06 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FK7
 
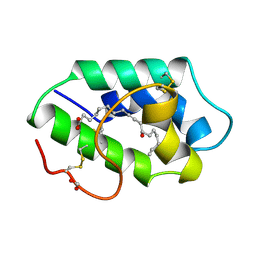 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH RICINOLEIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | 分子名称: | FORMIC ACID, NON-SPECIFIC LIPID TRANSFER PROTEIN, RICINOLEIC ACID | | 著者 | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | 登録日 | 2000-08-09 | | 公開日 | 2001-06-06 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FK3
 
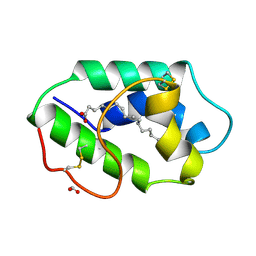 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH PALMITOLEIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | 分子名称: | FORMIC ACID, NONSPECIFIC LIPID-TRANSFER PROTEIN, PALMITOLEIC ACID | | 著者 | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | 登録日 | 2000-08-09 | | 公開日 | 2001-06-06 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FK0
 
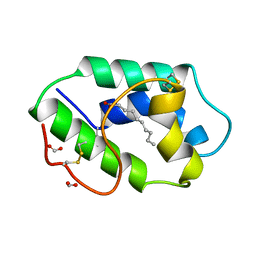 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH CAPRIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | 分子名称: | DECANOIC ACID, FORMIC ACID, NONSPECIFIC LIPID-TRANSFER PROTEIN | | 著者 | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | 登録日 | 2000-08-08 | | 公開日 | 2001-06-06 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
4FBB
 
 | | Structure of mutant RIP from barley seeds in complex with adenine (AMP-incubated) | | 分子名称: | ADENINE, Protein synthesis inhibitor I | | 著者 | Lee, B.-G, Kim, M.K, Suh, S.W, Song, H.K. | | 登録日 | 2012-05-22 | | 公開日 | 2012-10-31 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structures of the ribosome-inactivating protein from barley seeds reveal a unique activation mechanism.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4FBH
 
 | | Structure of RIP from barley seeds | | 分子名称: | ADENOSINE MONOPHOSPHATE, Protein synthesis inhibitor I | | 著者 | Lee, B.-G, Kim, M.K, Suh, S.W, Song, H.K. | | 登録日 | 2012-05-23 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structures of the ribosome-inactivating protein from barley seeds reveal a unique activation mechanism.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4FBC
 
 | | Structure of mutant RIP from barley seeds in complex with AMP | | 分子名称: | ADENOSINE MONOPHOSPHATE, Protein synthesis inhibitor I | | 著者 | Lee, B.-G, Kim, M.K, Suh, S.W, Song, H.K. | | 登録日 | 2012-05-22 | | 公開日 | 2012-10-31 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structures of the ribosome-inactivating protein from barley seeds reveal a unique activation mechanism.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4FB9
 
 | | Structure of mutant RIP from barley seeds | | 分子名称: | Protein synthesis inhibitor I | | 著者 | Lee, B.-G, Kim, M.K, Suh, S.W, Song, H.K. | | 登録日 | 2012-05-22 | | 公開日 | 2012-10-31 | | 最終更新日 | 2013-01-23 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structures of the ribosome-inactivating protein from barley seeds reveal a unique activation mechanism.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3TT7
 
 | |
3TT6
 
 | |
5IZV
 
 | | Crystal structure of the legionella pneumophila effector protein RavZ - F222 | | 分子名称: | Uncharacterized protein RavZ | | 著者 | Kwon, D.H, Kim, L, Kim, B.-W, Hong, S.B, Song, H.K. | | 登録日 | 2016-03-26 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.814 Å) | | 主引用文献 | The 1:2 complex between RavZ and LC3 reveals a mechanism for deconjugation of LC3 on the phagophore membrane
Autophagy, 13, 2017
|
|
5XUY
 
 | | Crystal structure of ATG101-ATG13HORMA | | 分子名称: | Autophagy-related protein 101, Autophagy-related protein 13 | | 著者 | Kim, B.-W, Song, H.K. | | 登録日 | 2017-06-26 | | 公開日 | 2018-07-04 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The C-terminal region of ATG101 bridges ULK1 and PtdIns3K complex in autophagy initiation.
Autophagy, 14, 2018
|
|
5XAC
 
 | | CLIR - LC3B | | 分子名称: | Microtubule-associated proteins 1A/1B light chain 3B | | 著者 | Kwon, D.H, Kim, L, Song, H.K. | | 登録日 | 2017-03-12 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.701 Å) | | 主引用文献 | A novel conformation of the LC3-interacting region motif revealed by the structure of a complex between LC3B and RavZ
Biochem. Biophys. Res. Commun., 490, 2017
|
|
7YRB
 
 | | UBR box of human UBR6 | | 分子名称: | F-box protein 11, isoform CRA_f, SULFATE ION, ... | | 著者 | Kim, B, Song, H.K. | | 登録日 | 2022-08-09 | | 公開日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Crystal structure of UBR box from human UBR6
To Be Published
|
|
7P80
 
 | | Crystal structure of ClpP from Bacillus subtilis in complex with ADEP2 (compressed state) | | 分子名称: | ADEP2, ATP-dependent Clp protease proteolytic subunit | | 著者 | Lee, B.-G, Kim, L, Kim, M.K, Kwon, D.H, Song, H.K. | | 登録日 | 2021-07-21 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | Structural insights into ClpP protease side exit pore-opening by a pH drop coupled with substrate hydrolysis.
Embo J., 41, 2022
|
|
7P81
 
 | | Crystal structure of ClpP from Bacillus subtilis in complex with ADEP2 (compact state) | | 分子名称: | ADEP2, ATP-dependent Clp protease proteolytic subunit | | 著者 | Lee, B.-G, Kim, L, Kim, M.K, Kwon, D.H, Song, H.K. | | 登録日 | 2021-07-21 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Structural insights into ClpP protease side exit pore-opening by a pH drop coupled with substrate hydrolysis.
Embo J., 41, 2022
|
|
3KTJ
 
 | |
3KTI
 
 | | Structure of ClpP in complex with ADEP1 | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ATP-dependent Clp protease proteolytic subunit, Acyldepsipeptide 1, ... | | 著者 | Lee, B.-G, Brotz-Oesterhelt, H, Song, H.K. | | 登録日 | 2009-11-25 | | 公開日 | 2010-03-23 | | 最終更新日 | 2013-02-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structures of ClpP in complex with acyldepsipeptide antibiotics reveal its activation mechanism
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KTK
 
 | |
3KTG
 
 | |
3KTH
 
 | |
4EBR
 
 | | Crystal structure of Autophagic E2, Atg10 | | 分子名称: | MERCURY (II) ION, Ubiquitin-like-conjugating enzyme ATG10 | | 著者 | Hong, S.B, Kim, B.W, Kim, J.H, Song, H.K. | | 登録日 | 2012-03-24 | | 公開日 | 2012-10-03 | | 最終更新日 | 2013-07-24 | | 実験手法 | X-RAY DIFFRACTION (2.701 Å) | | 主引用文献 | Structure of the autophagic E2 enzyme Atg10
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6L18
 
 | |
6LHN
 
 | | RLGSGG-AtPRT6 UBR box | | 分子名称: | E3 ubiquitin-protein ligase PRT6, ZINC ION | | 著者 | Kim, L, Kwon, D.H, Song, H.K. | | 登録日 | 2019-12-09 | | 公開日 | 2020-01-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Use of the LC3B-fusion technique for biochemical and structural studies of proteins involved in the N-degron pathway.
J.Biol.Chem., 295, 2020
|
|
1V9P
 
 | | Crystal Structure Of Nad+-Dependent DNA Ligase | | 分子名称: | ADENOSINE MONOPHOSPHATE, DNA ligase, ZINC ION | | 著者 | Lee, J.Y, Chang, C, Song, H.K, Moon, J, Yang, J.K, Kim, H.K, Kwon, S.K, Suh, S.W. | | 登録日 | 2004-01-27 | | 公開日 | 2004-03-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of NAD(+)-dependent DNA ligase: modular architecture and functional implications.
Embo J., 19, 2000
|
|
