8Q88
 
 | |
8Q2K
 
 | | Tau - AD-MIA3 | | 分子名称: | Isoform Tau-D of Microtubule-associated protein tau | | 著者 | Lovestam, S, Li, D, Scheres, S.H.W, Goedert, M. | | 登録日 | 2023-08-02 | | 公開日 | 2023-08-30 | | 最終更新日 | 2024-01-24 | | 実験手法 | ELECTRON MICROSCOPY (2.88 Å) | | 主引用文献 | Disease-specific tau filaments assemble via polymorphic intermediates.
Nature, 625, 2024
|
|
8Q7P
 
 | |
8Q7T
 
 | | Tau - AD-MIA11 | | 分子名称: | Isoform Tau-D of Microtubule-associated protein tau | | 著者 | Lovestam, S, Li, D, Scheres, S.H.W, Goedert, M. | | 登録日 | 2023-08-17 | | 公開日 | 2023-08-30 | | 最終更新日 | 2024-01-24 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Disease-specific tau filaments assemble via polymorphic intermediates.
Nature, 625, 2024
|
|
8QCP
 
 | |
8Q9I
 
 | |
8Q9O
 
 | |
8Q8M
 
 | |
8Q98
 
 | |
8DM4
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein in complex with Fab 4A8 (focused refinement of NTD and 4A8) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 4A8 heavy chain, ... | | 著者 | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | 登録日 | 2022-07-08 | | 公開日 | 2023-01-25 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (2.45 Å) | | 主引用文献 | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
8DM6
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein in complex with human ACE2 (focused refinement of RBD and ACE2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | 著者 | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | 登録日 | 2022-07-08 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (2.77 Å) | | 主引用文献 | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
8DMA
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.1 spike protein in complex with mouse ACE2 (focused refinement of RBD and ACE2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein | | 著者 | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | 登録日 | 2022-07-08 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (2.79 Å) | | 主引用文献 | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
4QI4
 
 | | Dehydrogenase domain of Myriococcum thermophilum cellobiose dehydrogenase, MtDH | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | 著者 | Tan, T.C, Gandini, R, Sygmund, C, Kittl, R, Haltrich, D, Ludwig, R, Hallberg, B.M, Divne, C. | | 登録日 | 2014-05-30 | | 公開日 | 2015-07-15 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural basis for cellobiose dehydrogenase action during oxidative cellulose degradation.
Nat Commun, 6, 2015
|
|
6RNX
 
 | | Crystal structure of the essential repressor DdrO from radiation-resistant Deinococcus bacteria (Deinococcus deserti) | | 分子名称: | CHLORIDE ION, HTH-type transcriptional regulator DdrOC | | 著者 | Arnoux, P, Siponen, M.I, Pignol, D, De Groot, A, Blanchard, L. | | 登録日 | 2019-05-09 | | 公開日 | 2019-10-09 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.84 Å) | | 主引用文献 | Crystal structure of the transcriptional repressor DdrO: insight into the metalloprotease/repressor-controlled radiation response in Deinococcus.
Nucleic Acids Res., 47, 2019
|
|
8Q2L
 
 | | Tau - AD-MIA4 | | 分子名称: | Isoform Tau-D of Microtubule-associated protein tau | | 著者 | Lovestam, S, Li, D, Scheres, S.H.W, Goedert, M. | | 登録日 | 2023-08-02 | | 公開日 | 2023-09-20 | | 最終更新日 | 2024-01-24 | | 実験手法 | ELECTRON MICROSCOPY (2.2 Å) | | 主引用文献 | Disease-specific tau filaments assemble via polymorphic intermediates.
Nature, 625, 2024
|
|
8Q97
 
 | |
8QCR
 
 | |
8DM7
 
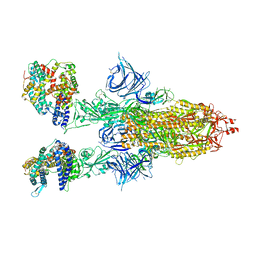 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein in complex with mouse ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | 登録日 | 2022-07-08 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (2.49 Å) | | 主引用文献 | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
8Q9D
 
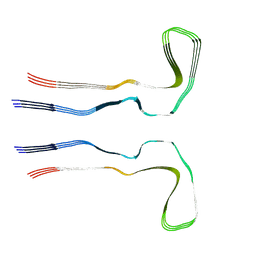 | |
8DM5
 
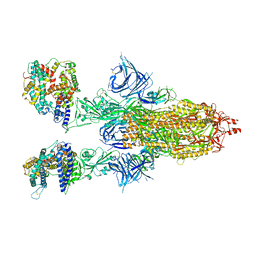 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein in complex with human ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | 登録日 | 2022-07-08 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (2.51 Å) | | 主引用文献 | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
8Q8R
 
 | | Tau - AD-PHF | | 分子名称: | Isoform Tau-D of Microtubule-associated protein tau | | 著者 | Lovestam, S, Li, D, Scheres, S.H.W, Goedert, M. | | 登録日 | 2023-08-18 | | 公開日 | 2023-09-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (2.1 Å) | | 主引用文献 | Disease-specific tau filaments assemble via polymorphic intermediates.
Nature, 625, 2024
|
|
8Q99
 
 | |
8Q9H
 
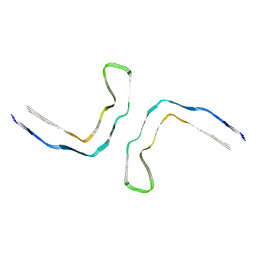 | |
8Q8F
 
 | |
8DM8
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein in complex with mouse ACE2 (focused refinement of RBD and ACE2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein | | 著者 | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | 登録日 | 2022-07-08 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (2.68 Å) | | 主引用文献 | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
