3V4E
 
 | | Crystal Structure of the galactoside O-acetyltransferase in complex with CoA | | 分子名称: | COENZYME A, DI(HYDROXYETHYL)ETHER, Galactoside O-acetyltransferase, ... | | 著者 | Knapik, A.A, Shumilin, I.A, Luo, H.-B, Chruszcz, M, Zimmerman, M.D, Cymborowski, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-12-14 | | 公開日 | 2012-01-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Biophysical analysis of the putative acetyltransferase SACOL2570 from methicillin-resistant Staphylococcus aureus.
J.Struct.Funct.Genom., 14, 2013
|
|
3UEE
 
 | | Crystal structure of human Survivin K62A mutant bound to N-terminal histone H3 | | 分子名称: | Baculoviral IAP repeat-containing protein 5, N-terminal fragment of histone H3, ZINC ION | | 著者 | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | 登録日 | 2011-10-30 | | 公開日 | 2012-03-07 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
3V08
 
 | | Crystal structure of Equine Serum Albumin | | 分子名称: | 1,2-ETHANEDIOL, BROMIDE ION, SULFATE ION, ... | | 著者 | Dayal, A, Jablonska, K, Porebski, P.J, Majorek, K.A, Chruszcz, M, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2011-12-07 | | 公開日 | 2012-01-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structural and immunologic characterization of bovine, horse, and rabbit serum albumins.
Mol.Immunol., 52, 2012
|
|
3TZK
 
 | | Crystal structure of 3-ketoacyl-(acyl-carrier-protein) reductase (FabG)(G92A) from Vibrio cholerae | | 分子名称: | 3-oxoacyl-[acyl-carrier protein] reductase, SULFATE ION, UNKNOWN ATOM OR ION | | 著者 | Hou, J, Chruszcz, M, Zheng, H, Grabowski, M, Domagalski, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-09-27 | | 公開日 | 2011-10-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Dissecting the Structural Elements for the Activation of beta-Ketoacyl-(Acyl Carrier Protein) Reductase from Vibrio cholerae.
J.Bacteriol., 198, 2015
|
|
3TNJ
 
 | | Crystal structure of universal stress protein from Nitrosomonas europaea with AMP bound | | 分子名称: | ADENOSINE MONOPHOSPHATE, Universal stress protein (Usp) | | 著者 | Tkaczuk, K.L, Chruszcz, M, Shumilin, I.A, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-09-01 | | 公開日 | 2011-09-14 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and functional insight into the universal stress protein family.
Evol Appl, 6, 2013
|
|
3TPF
 
 | | Crystal structure of anabolic ornithine carbamoyltransferase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | 分子名称: | DI(HYDROXYETHYL)ETHER, Ornithine carbamoyltransferase | | 著者 | Shabalin, I.G, Onopriyenko, O, Grimshaw, S, Porebski, P.J, Grabowski, M, Savchenko, A, Chruszcz, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-09-07 | | 公開日 | 2011-09-21 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure of anabolic ornithine carbamoyltransferase from Campylobacter jejuni at 2.7 A resolution.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3UDU
 
 | | Crystal structure of putative 3-isopropylmalate dehydrogenase from Campylobacter jejuni | | 分子名称: | 1,2-ETHANEDIOL, 3-isopropylmalate dehydrogenase, CHLORIDE ION | | 著者 | Tkaczuk, K.L, Chruszcz, M, Grimshaw, S, Onopriyenko, O, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-10-28 | | 公開日 | 2011-11-09 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structure of putative 3-isopropylmalate dehydrogenase from Campylobacter jejuni
To be Published
|
|
3UEG
 
 | | Crystal structure of human Survivin K62A mutant | | 分子名称: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 5, TETRAETHYLENE GLYCOL, ... | | 著者 | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | 登録日 | 2011-10-30 | | 公開日 | 2012-03-07 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
3UEC
 
 | | Crystal structure of human Survivin bound to histone H3 phosphorylated on threonine-3. | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, Baculoviral IAP repeat-containing protein 5, ... | | 著者 | Niedzialkowska, E, Porebski, P.J, Cooper, D.R, Chruszcz, M, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | 登録日 | 2011-10-30 | | 公開日 | 2012-03-07 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
3UEI
 
 | | Crystal structure of human Survivin E65A mutant | | 分子名称: | Baculoviral IAP repeat-containing protein 5, ZINC ION | | 著者 | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | 登録日 | 2011-10-30 | | 公開日 | 2012-03-07 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol Biol Cell, 23, 2012
|
|
3V0R
 
 | | Crystal structure of Alternaria alternata allergen Alt a 1 | | 分子名称: | 2,5,6-triaminopyrimidin-4-ol, 8-aminooctanoic acid, Major allergen Alt a 1, ... | | 著者 | Chruszcz, M, Solberg, R, Osinski, T, Chapman, M.D, Minor, W. | | 登録日 | 2011-12-08 | | 公開日 | 2012-06-13 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Alternaria alternata allergen Alt a 1: a unique beta-barrel protein dimer found exclusively in fungi.
J.Allergy Clin.Immunol., 130, 2012
|
|
3V48
 
 | | Crystal Structure of the putative alpha/beta hydrolase RutD from E.coli | | 分子名称: | GLYCEROL, Putative aminoacrylate hydrolase RutD, THIOCYANATE ION | | 著者 | Knapik, A.A, Petkowski, J.J, Otwinowski, Z, Cymborowski, M.T, Cooper, D.R, Chruszcz, M, Porebski, P.J, Niedzialkowska, E, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2011-12-14 | | 公開日 | 2012-01-04 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A multi-faceted analysis of RutD reveals a novel family of alpha / beta hydrolases.
Proteins, 80, 2012
|
|
3V4D
 
 | | Crystal structure of RutC protein a member of the YjgF family from E.coli | | 分子名称: | Aminoacrylate peracid reductase RutC | | 著者 | Knapik, A.A, Petkowski, J.J, Otwinowski, Z, Cymborowski, M.T, Cooper, D.R, Chruszcz, M, Porebski, P.J, Niedzialkowska, E, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2011-12-14 | | 公開日 | 2012-01-04 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure of Escherichia coli RutC, a member of the YjgF family and putative aminoacrylate peracid reductase of the rut operon.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3V03
 
 | | Crystal structure of Bovine Serum Albumin | | 分子名称: | ACETATE ION, CALCIUM ION, Serum albumin | | 著者 | Majorek, K.A, Porebski, P.J, Chruszcz, M, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2011-12-07 | | 公開日 | 2012-01-04 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural and immunologic characterization of bovine, horse, and rabbit serum albumins.
Mol.Immunol., 52, 2012
|
|
5O2E
 
 | | Crystal structure of NDM-1 in complex with hydrolyzed cefuroxime - new refinement | | 分子名称: | (2R,5S)-5-[(carbamoyloxy)methyl]-2-[(R)-carboxy{[(2Z)-2-(furan-2-yl)-2-(methoxyimino)acetyl]amino}methyl]-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Metallo-beta-lactamase type 2, SULFATE ION, ... | | 著者 | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A. | | 登録日 | 2017-05-20 | | 公開日 | 2018-12-26 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
5O2F
 
 | | Crystal structure of NDM-1 in complex with hydrolyzed ampicillin - new refinement | | 分子名称: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | 著者 | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A. | | 登録日 | 2017-05-20 | | 公開日 | 2018-12-26 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
1VCP
 
 | |
6OV8
 
 | | 2.6 Angstrom Resolution Crystal Structure of Aminopeptidase B from Escherichia coli str. K-12 substr. MG1655 | | 分子名称: | CHLORIDE ION, MANGANESE (II) ION, Peptidase B, ... | | 著者 | Minasov, G, Shuvalova, L, Wawrzak, Z, Kiryukhina, O, Grimshaw, S, Kwon, K, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-05-07 | | 公開日 | 2019-05-15 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Comparison of metal-bound and unbound structures of aminopeptidase B proteins from Escherichia coli and Yersinia pestis.
Protein Sci., 29, 2020
|
|
6OAD
 
 | | 2.05 Angstrom Resolution Crystal Structure of Aminopeptidase B from Escherichia coli str. K-12 substr. MG1655. | | 分子名称: | 1,2-ETHANEDIOL, BICARBONATE ION, CALCIUM ION, ... | | 著者 | Minasov, G, Shuvalova, L, Wawrzak, Z, Kiryukhina, O, Grimshaw, S, Kwon, K, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-03-15 | | 公開日 | 2019-03-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Comparison of metal-bound and unbound structures of aminopeptidase B proteins from Escherichia coli and Yersinia pestis.
Protein Sci., 29, 2020
|
|
7P18
 
 | | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Sterolibacterium denitrificans in complex with 1,4-androstadiene-3,17-dione | | 分子名称: | 3-oxosteroid 1-dehydrogenase, ANDROSTA-1,4-DIENE-3,17-DIONE, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Wojcik, P, Mrugala, B, Kurpiewska, K, Szaleniec, M. | | 登録日 | 2021-07-01 | | 公開日 | 2021-07-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Structure, Mutagenesis, and QM:MM Modeling of 3-Ketosteroid Delta 1 -Dehydrogenase from Sterolibacterium denitrificans ─The Role of a New Putative Membrane-Associated Domain and Proton-Relay System in Catalysis.
Biochemistry, 62, 2023
|
|
2SNV
 
 | |
3LNL
 
 | | Crystal structure of Staphylococcus aureus protein SA1388 | | 分子名称: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, UPF0135 protein SA1388, ZINC ION | | 著者 | Singh, K.S, Chruszcz, M, Zhang, X, Minor, W, Zhang, H. | | 登録日 | 2010-02-02 | | 公開日 | 2010-03-16 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of a Conserved Hypothetical Protein Sa1388 from S. aureus Reveals a Capped Hexameric Toroid with Two Pii Domain Lids and a Dinuclear Metal Center.
Bmc Struct.Biol., 6, 2006
|
|
2RD7
 
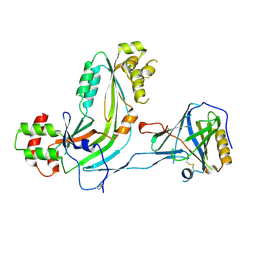 | | Human Complement Membrane Attack Proteins Share a Common Fold with Bacterial Cytolysins | | 分子名称: | CHLORIDE ION, Complement component C8 alpha chain, Complement component C8 gamma chain | | 著者 | Slade, D.J, Lovelace, L.L, Chruszcz, M, Minor, W, Lebioda, L, Sodetz, J.M. | | 登録日 | 2007-09-21 | | 公開日 | 2008-05-27 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structure of human C8 protein provides mechanistic insight into membrane pore formation by complement.
J. Biol. Chem., 286, 2011
|
|
3LOC
 
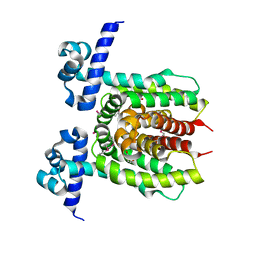 | | Crystal structure of putative transcriptional regulator ycdc | | 分子名称: | HTH-type transcriptional regulator rutR, URACIL | | 著者 | Patskovsky, Y.V, Knapik, A.A, Mennella, V, Burley, S.K, Minor, W, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2010-02-03 | | 公開日 | 2010-03-16 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of Hypothetical Transcriptional Regulator Ycdc from Escherichia Coli
To be Published
|
|
2SNW
 
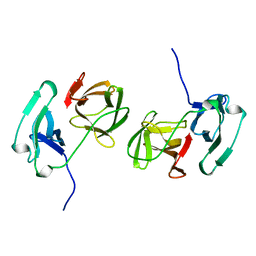 | | SINDBIS VIRUS CAPSID PROTEIN, TYPE3 CRYSTAL FORM | | 分子名称: | COAT PROTEIN C | | 著者 | Choi, H.-K, Lee, S, Zhang, Y.-P, Mckinney, B.R, Wengler, G, Rossmann, M.G, Kuhn, R.J. | | 登録日 | 1998-02-17 | | 公開日 | 1998-04-08 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural analysis of Sindbis virus capsid mutants involving assembly and catalysis.
J.Mol.Biol., 262, 1996
|
|
