8GX2
 
 | | The crystal structure of human CtsL in complex with 14c | | 分子名称: | DIMETHYL SULFOXIDE, N-[(2S)-3-cyclohexyl-1-[[(2S,3S)-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-1-benzofuran-2-carboxamide, Procathepsin L | | 著者 | Zhao, Y, Shao, M, Zhao, J, Yang, H, Rao, Z. | | 登録日 | 2022-09-18 | | 公開日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The crystal structure of human CtsL in complex with 14a
To Be Published
|
|
8GXG
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with 14a | | 分子名称: | 3C-like proteinase nsp5, N-[(2S)-3-(4-fluorophenyl)-1-oxidanylidene-1-[[(2S,3S)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide | | 著者 | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | 登録日 | 2022-09-20 | | 公開日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | The crystal structure of SARS-CoV-2 main protease in complex with 14a
To Be Published
|
|
8GXH
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with 14b | | 分子名称: | 3C-like proteinase nsp5, N-[(2S)-3-cyclohexyl-1-oxidanylidene-1-[[(2S,3R)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide | | 著者 | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | 登録日 | 2022-09-20 | | 公開日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | The crystal structure of SARS-CoV-2 main protease in complex with 14b
To Be Published
|
|
8H68
 
 | | Crystal structure of Caenorhabditis elegans NMAD-1 in complex with NOG and Mg(II) | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Shi, Y, Ding, J, Yang, H. | | 登録日 | 2022-10-16 | | 公開日 | 2023-02-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Caenorhabditis elegans NMAD-1 functions as a demethylase for actin.
J Mol Cell Biol, 15, 2023
|
|
8HNQ
 
 | | The structure of a alcohol dehydrogenase AKR13B2 with NADP | | 分子名称: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Chen, M, Yang, H, Lu, F. | | 登録日 | 2022-12-08 | | 公開日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The structure of AKR13B2
To Be Published
|
|
7DVW
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp5|6 peptidyl substrate | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, nsp5/6 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7E5X
 
 | |
7DVP
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp4|5 peptidyl substrate | | 分子名称: | 3C-like proteinase, nsp4/5 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-14 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DW0
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp14|15 peptidyl substrate | | 分子名称: | 3C-like proteinase, nsp14/15 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DW6
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp15|16 peptidyl substrate | | 分子名称: | 3C-like proteinase, nsp15/16 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVY
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp9|10 peptidyl substrate | | 分子名称: | 3C-like proteinase, nsp9/10 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVX
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp6|7 peptidyl substrate | | 分子名称: | 3C-like proteinase, nsp6/7 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VMU
 
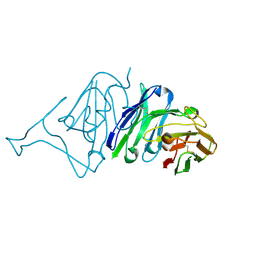 | | Crystal Structure of SARS-CoV Spike Receptor-Binding Domain Complexed with Neutralizing Antibody | | 分子名称: | Spike protein S1, scFv E4 | | 著者 | Guo, Y, Wang, W, Jiao, P, Yang, H, Rao, Z, Cheng, G. | | 登録日 | 2021-10-09 | | 公開日 | 2021-11-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Antibody engineering improves neutralization activity against K417 spike mutant SARS-CoV-2 variants.
Cell Biosci, 12, 2022
|
|
7VBA
 
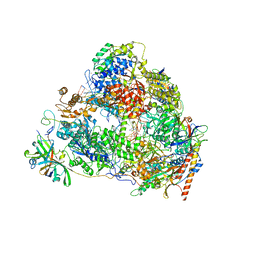 | | Structure of the pre state human RNA Polymerase I Elongation Complex | | 分子名称: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, DNA (5'-D(P*A*CP*TP*GP*TP*CP*CP*TP*CP*TP*GP*GP*C)-3'), DNA (5'-D(P*GP*CP*CP*AP*GP*AP*GP*AP*CP*AP*GP*CP*GP*AP*GP*TP*CP*AP*GP*CP*AP*A)-3'), ... | | 著者 | Zhao, D, Liu, W, Chen, K, Yang, H, Xu, Y. | | 登録日 | 2021-08-31 | | 公開日 | 2022-02-16 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.89 Å) | | 主引用文献 | Structure of the human RNA polymerase I elongation complex.
Cell Discov, 7, 2021
|
|
7VBC
 
 | | Back track state of human RNA Polymerase I Elongation Complex | | 分子名称: | DNA (5'-D(*GP*TP*AP*CP*TP*GP*TP*CP*CP*TP*CP*TP*GP*G)-3'), DNA (5'-D(P*AP*GP*GP*AP*CP*AP*GP*CP*GP*TP*GP*TP*CP*AP*GP*CP*AP*AP*TP*A)-3'), DNA-directed RNA polymerase I subunit RPA1, ... | | 著者 | Zhao, D, Liu, W, Chen, K, Yang, H, Xu, Y. | | 登録日 | 2021-08-31 | | 公開日 | 2022-02-16 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.01 Å) | | 主引用文献 | Structure of the human RNA polymerase I elongation complex.
Cell Discov, 7, 2021
|
|
7VBB
 
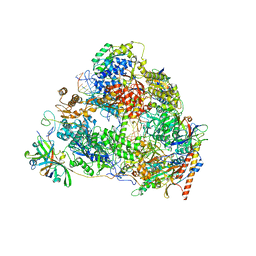 | | Structure of the post state human RNA Polymerase I Elongation Complex | | 分子名称: | DNA (25-MER), DNA (5'-D(*CP*TP*GP*TP*CP*CP*TP*CP*TP*GP*GP*CP*GP*A)-3'), DNA-directed RNA polymerase I subunit RPA1, ... | | 著者 | Zhao, D, Liu, W, Chen, K, Yang, H, Xu, Y. | | 登録日 | 2021-08-31 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.81 Å) | | 主引用文献 | Structure of the human RNA polymerase I elongation complex.
Cell Discov, 7, 2021
|
|
5ZHY
 
 | | Structural characterization of the HCoV-229E fusion core | | 分子名称: | Spike glycoprotein | | 著者 | Zhang, W, Zheng, Q, Yan, M, Chen, X, Yang, H, Zhou, W, Rao, Z. | | 登録日 | 2018-03-13 | | 公開日 | 2018-06-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.441 Å) | | 主引用文献 | Structural characterization of the HCoV-229E fusion core.
Biochem. Biophys. Res. Commun., 497, 2018
|
|
7WWK
 
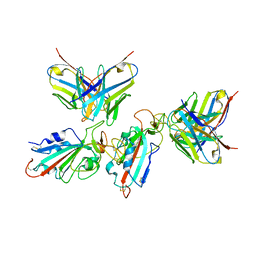 | | Local refinement of the SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab | | 分子名称: | 55A8 light chain, Spike glycoprotein | | 著者 | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | 登録日 | 2022-02-13 | | 公開日 | 2023-02-15 | | 最終更新日 | 2023-04-19 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Local refinement of the SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab
To Be Published
|
|
7WWJ
 
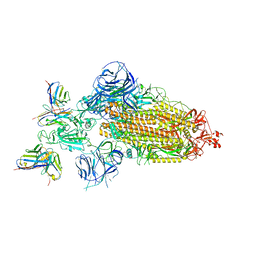 | | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab in the class 2 conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 55A8 heavy chain, ... | | 著者 | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | 登録日 | 2022-02-13 | | 公開日 | 2023-02-15 | | 最終更新日 | 2023-04-19 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | EM structure of SARS-CoV-2 Omicron variant spike glycoprotein and 55A8
To Be Published
|
|
7WWI
 
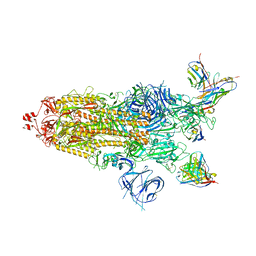 | | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab in the class 1 conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 55A8 heavy chain, ... | | 著者 | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | 登録日 | 2022-02-13 | | 公開日 | 2023-02-15 | | 最終更新日 | 2023-04-19 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab in the class 1 conformation
To Be Published
|
|
6B44
 
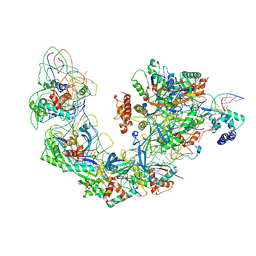 | | Cryo-EM structure of Type I-F CRISPR crRNA-guided Csy surveillance complex with bound target dsDNA | | 分子名称: | CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, CRISPR-associated protein Csy2, ... | | 著者 | Guo, T.W, Bartesaghi, A, Yang, H, Falconieri, V, Rao, P, Merk, A, Fox, T, Earl, L, Patel, D.J, Subramaniam, S. | | 登録日 | 2017-09-25 | | 公開日 | 2017-10-18 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
7XJ8
 
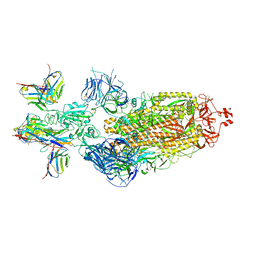 | | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab and 58G6 Fab in the class 2 conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 55A8 heavy chain, ... | | 著者 | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | 登録日 | 2022-04-15 | | 公開日 | 2023-04-19 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab and 58G6 Fab in the class 2 conformation
To Be Published
|
|
7XJ6
 
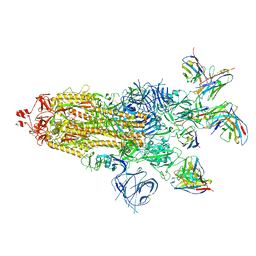 | | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab and 58G6 Fab in the class 1 conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 55A8 heavy chain, ... | | 著者 | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | 登録日 | 2022-04-15 | | 公開日 | 2023-04-19 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab and 58G6 Fab in the class 1 conformation
To Be Published
|
|
6B48
 
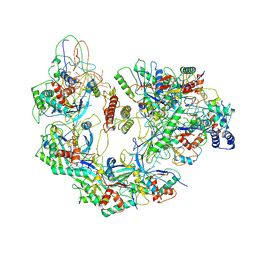 | | Cryo-EM structure of Type I-F CRISPR crRNA-guided Csy surveillance complex with bound anti-CRISPR protein AcrF10 | | 分子名称: | Anti-CRISPR protein AcrF10, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | 著者 | Guo, T.W, Bartesaghi, A, Yang, H, Falconieri, V, Rao, P, Merk, A, Fox, T, Earl, L, Patel, D.J, Subramaniam, S. | | 登録日 | 2017-09-25 | | 公開日 | 2017-10-18 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
6B47
 
 | | Cryo-EM structure of Type I-F CRISPR crRNA-guided Csy surveillance complex with bound anti-CRISPR protein AcrF2 | | 分子名称: | Anti-CRISPR protein AcrF2, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | 著者 | Guo, T.W, Bartesaghi, A, Yang, H, Falconieri, V, Rao, P, Merk, A, Fox, T, Earl, L, Patel, D.J, Subramaniam, S. | | 登録日 | 2017-09-25 | | 公開日 | 2017-10-18 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
