3G3F
 
 | |
3G3H
 
 | |
3G3K
 
 | |
3G3J
 
 | |
3G3G
 
 | |
3G3I
 
 | |
2XBJ
 
 | | Crystal Structure of Chk2 in complex with an inhibitor | | 分子名称: | 4-FLUORO-2-(4-{[(3S,4R)-4-(1-HYDROXY-1-METHYLETHYL)PYRROLIDIN-3-YL]AMINO}-6,7-DIMETHOXYQUINAZOLIN-2-YL)PHENOL, MAGNESIUM ION, NITRATE ION, ... | | 著者 | Anderson, V.E, Walton, M.I, Eve, P.D, Caldwell, J.J, Pearl, L.H, Oliver, A.W, Collins, I, Garrett, M.D. | | 登録日 | 2010-04-12 | | 公開日 | 2011-01-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure-Based Design of Potent and Selective 2-(Quinazolin-2-Yl)Phenol Inhibitors of Checkpoint Kinase 2.
J.Med.Chem., 54, 2011
|
|
2JX6
 
 | | Structure and membrane interactions of the antibiotic peptide dermadistinctin k by solution and oriented 15N and 31P solid-state NMR spectroscopy | | 分子名称: | Dermadistinctin-K | | 著者 | Mendonca Moraes, C, Verly, R.M, Resende, J.M, Bemquerer, M.P, Pilo-Veloso, D, Valente, A, Almeida, F.C.L, Bechinger, B. | | 登録日 | 2007-11-08 | | 公開日 | 2008-11-11 | | 最終更新日 | 2022-03-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and membrane interactions of the antibiotic peptide dermadistinctin K by multidimensional solution and oriented 15N and 31P solid-state NMR spectroscopy
Biophys.J., 96, 2009
|
|
1RRU
 
 | | The influence of a chiral amino acid on the helical handedness of PNA in solution and in crystals | | 分子名称: | Peptide Nucleic Acid, (H-P(*CPN*GPN*TPN*APN*CPN*GPN)-LYS-NH2) | | 著者 | Rasmussen, H, Liljefors, T, Petersson, B, Nielsen, P.E, Kastrup, J.S. | | 登録日 | 2003-12-09 | | 公開日 | 2004-01-13 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | The Influence of a Chiral Amino Acid on the Helical Handedness of PNA in Solution and in Crystals
J.Biomol.Struct.Dyn., 21, 2004
|
|
7KQV
 
 | |
7KRG
 
 | |
1XH1
 
 | | Structure of the N298S variant of human pancreatic alpha-amylase complexed with chloride | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, pancreatic, ... | | 著者 | Maurus, R, Begum, A, Kuo, H.H, Racaza, A, Numao, S, Overall, C.M, Withers, S.G, Brayer, G.D. | | 登録日 | 2004-09-17 | | 公開日 | 2005-05-24 | | 最終更新日 | 2021-11-10 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Structural and mechanistic studies of chloride induced activation of human pancreatic alpha-amylase
PROTEIN SCI., 14, 2005
|
|
1XH2
 
 | | Structure of the N298S variant of human pancreatic alpha-amylase complexed with chloride and acarbose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACARBOSE DERIVED PENTASACCHARIDE, Alpha-amylase, ... | | 著者 | Maurus, R, Begum, A, Kuo, H.H, Racaza, A, Numao, S, Overall, C.M, Withers, S.G, Brayer, G.D. | | 登録日 | 2004-09-17 | | 公開日 | 2005-05-24 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural and mechanistic studies of chloride induced activation of human pancreatic alpha-amylase
PROTEIN SCI., 14, 2005
|
|
2XJK
 
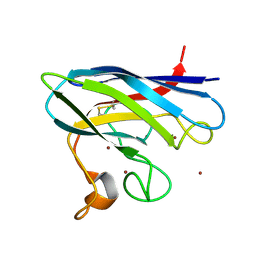 | | Monomeric Human Cu,Zn Superoxide dismutase | | 分子名称: | COPPER (II) ION, SUPEROXIDE DISMUTASE [CU-ZN], ZINC ION | | 著者 | Saraboji, K, Leinartaite, L, Nordlund, A, Oliveberg, M, Logan, D.T. | | 登録日 | 2010-07-07 | | 公開日 | 2010-09-01 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Folding Catalysis by Transient Coordination of Zn2+ to the Cu Ligands of the Als-Associated Enzyme Cu/Zn Superoxide Dismutase 1.
J.Am.Chem.Soc., 132, 2010
|
|
1XH0
 
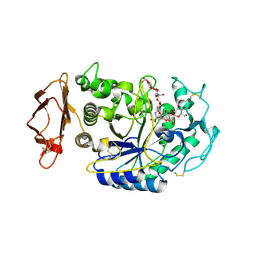 | | Structure of the N298S variant of human pancreatic alpha-amylase complexed with acarbose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACARBOSE DERIVED HEXASACCHARIDE, Alpha-amylase, ... | | 著者 | Maurus, R, Begum, A, Kuo, H.H, Racaza, A, Numao, S, Overall, C.M, Withers, S.G, Brayer, G.D. | | 登録日 | 2004-09-17 | | 公開日 | 2005-05-24 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and mechanistic studies of chloride induced activation of human pancreatic alpha-amylase
PROTEIN SCI., 14, 2005
|
|
1XGZ
 
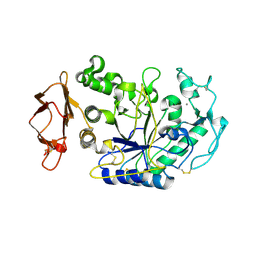 | | Structure of the N298S variant of human pancreatic alpha-amylase | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, pancreatic, ... | | 著者 | Maurus, R, Begum, A, Kuo, H.H, Racaza, A, Numao, S, Overall, C.M, Withers, S.G, Brayer, G.D. | | 登録日 | 2004-09-17 | | 公開日 | 2005-05-24 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and mechanistic studies of chloride induced activation of human pancreatic alpha-amylase
PROTEIN SCI., 14, 2005
|
|
1SO8
 
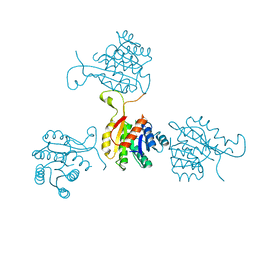 | | Abeta-bound human ABAD structure [also known as 3-hydroxyacyl-CoA dehydrogenase type II (Type II HADH), Endoplasmic reticulum-associated amyloid beta-peptide binding protein (ERAB)] | | 分子名称: | 3-hydroxyacyl-CoA dehydrogenase type II, CHLORIDE ION, SODIUM ION | | 著者 | Lustbader, J.W, Cirilli, M, Wu, H. | | 登録日 | 2004-03-13 | | 公開日 | 2004-05-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | ABAD directly links Abeta to mitochondrial toxicity in Alzheimer's disease.
Science, 304, 2004
|
|
3EXV
 
 | |
3F0Y
 
 | | Crystal structure of the human Adenovirus type 14 fiber knob | | 分子名称: | Fiber protein, GLYCEROL, IMIDAZOLE | | 著者 | Persson, B.D, Reiter, D.M, Arnberg, N, Stehle, T. | | 登録日 | 2008-10-27 | | 公開日 | 2008-11-25 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | An arginine switch in the species B adenovirus knob determines high-affinity engagement of cellular receptor CD46
J.Virol., 83, 2009
|
|
3F9I
 
 | |
3EXW
 
 | |
2GFE
 
 | |
1YKA
 
 | | Solution structure of Grx4, a monothiol glutaredoxin from E. coli. | | 分子名称: | monothiol glutaredoxin ydhD | | 著者 | Fladvad, M, Bellanda, M, Fernandes, A.P, Andresen, C, Mammi, S, Holmgren, A, Vlamis-Gardikas, A, Sunnerhagen, M. | | 登録日 | 2005-01-17 | | 公開日 | 2005-04-26 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Molecular mapping of functionalities in the solution structure of reduced Grx4, a monothiol glutaredoxin from Escherichia coli.
J.Biol.Chem., 280, 2005
|
|
2I0C
 
 | |
2I0B
 
 | |
